

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_204050 | SP | 0.140503 | 0.858877 | 0.000619 | CS pos: 23-24. ARG-LR. Pr: 0.6743 |
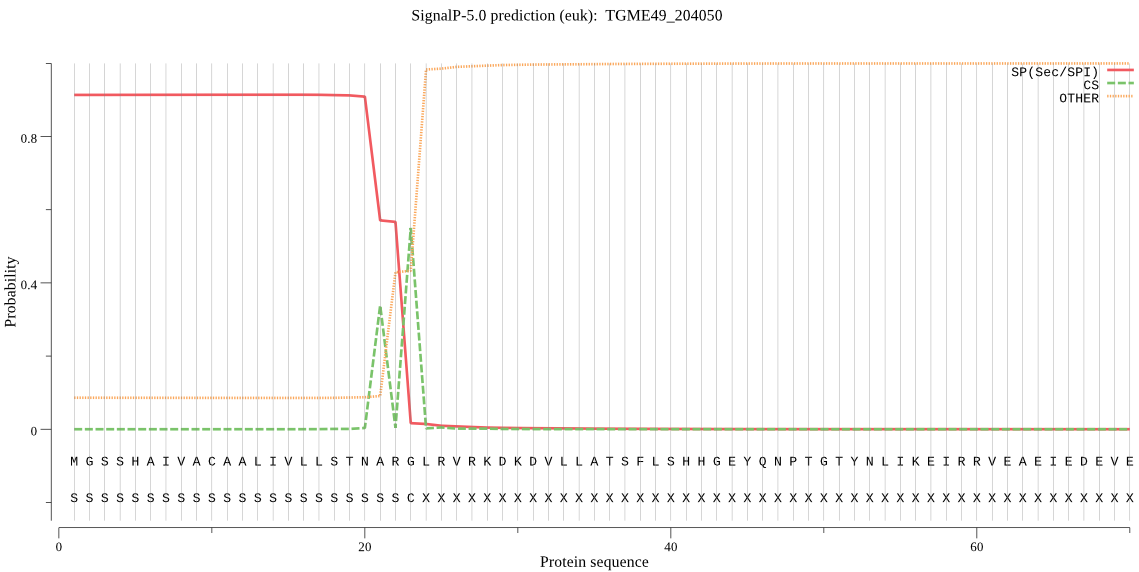
MGSSHAIVACAALIVLLSTNARGLRVRKDKDVLLATSFLSHHGEYQNPTGTYNLIKEIRR VEAEIEDEVETLNRDRRLHRGHNKYADDDIRQGLKGEQDMGASENIPVAELEPQDLDREA KYPVRMLIVDKRSDDDDEETKTSFVETALHSDLAQRVVKELNGHVDVLRESGVVLVDLPA QTTDKQLQELIETARAQGTIVEPDHLVQSVNTSSKGSNDPLLDRLWGMDALNVKEAWDII TTGEPNMGGRRPLVCVLDTGIDYNHPDLRDNMEVNQAERDGTLGVDDDNNGEVDDIYGAN MLSKENDPADDHSHGTHVAGTIGAHGNNGIGVAGVAWAPRLLPCKFLAYTGRGYSSDAVR CIDYCVKRGADIVNHSWGGSWPSEALREAVVRTANNGLIHIFAAGNDGVDIDQTAFYPAA FSTEADGLITVANVKGDPDHGGKRIIELDRSSNYGIQRVQVACPGMWILSTVPTSGSSQE PYAEKSGTSMAAPALSGIVALMLAVNPGLSTRQVREGLRQCSVQQPLLQGKVEWGSMPDA KRCVEYALTTHAEGRHKSFIREPSTETSTPPPPPPAQPTPQPQPHPPPQPETPPCAPSPP PPTPPCAPSPSPPTPPPGSPHKPEPQTPVYPEVPRSTRSPPPSPPPTESAPEAPPSDTPS CRVPPCSSSPRSGSQPKPPQDNTTTPKMPSLSSPPTEHSTAQPPKHENDAREQEPPTDED DFSSVKGKKLGAYESDGSPRASSCAGAGVLGVFFMVVGLTV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_204050 | 564 S | IREPSTETS | 0.991 | unsp | TGME49_204050 | 564 S | IREPSTETS | 0.991 | unsp | TGME49_204050 | 564 S | IREPSTETS | 0.991 | unsp | TGME49_204050 | 619 S | PPPGSPHKP | 0.991 | unsp | TGME49_204050 | 636 S | EVPRSTRSP | 0.993 | unsp | TGME49_204050 | 643 S | SPPPSPPPT | 0.996 | unsp | TGME49_204050 | 672 S | SSPRSGSQP | 0.993 | unsp | TGME49_204050 | 738 S | ESDGSPRAS | 0.994 | unsp | TGME49_204050 | 133 S | VDKRSDDDD | 0.997 | unsp | TGME49_204050 | 475 S | TVPTSGSSQ | 0.993 | unsp |