

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
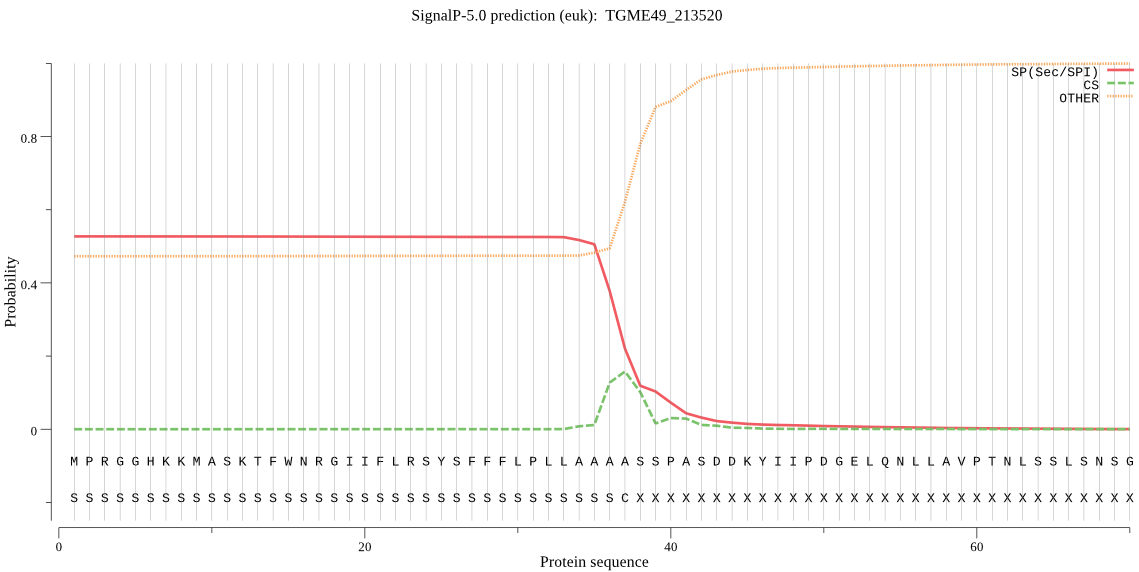
| TGME49_213520 | SP | 0.246635 | 0.744164 | 0.009201 | CS pos: 38-39. AAS-SP. Pr: 0.3411 |
MPRGGHKKMASKTFWNRGIIFLRSYSFFFLPLLAAAASSPASDDKYIIPDGELQNLLAVP TNLSSLSNSGSLVDFSTLFEAASETSFNAWIVAVRRALHQWPETAYNEYRTSALIHKLLK AMNVRVTTGWGTNTIGMSEEEAKIARARREGTGLVAEIGTGKEPCVALRADIDALPIFER TNVPFRSKVDGQMHACGHDVHTTMLLGAAALLKQLEPHMEGTIRLIFQPAEEGGGGALMM REEGVLTMAPPVEFIFGMHVAPALPTGELATRKGAMMAAATQFSINVKGRGGHGAVPHET IDPSPGVAAIVQGLYAIVARETSFTENTTGLISVTRIQGGTAFNVIPSEYFIGGTIRALD MAMMRNLQARVVELVENLAQAFRCQADVKYGSVSYVPLVNDPDATEFFIQTAAPASRSGR VGIADPTLGGEDFAFFLEDVPGTFAVIGIGSGAEHQLGHVPTNIPLHNPNFAVDERVLNR GAAVHAFTALRAFSFLASKKRNAGAAETRTTCEA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_213520 | 46 Y | SDDKYIIPD | 0.992 | unsp | TGME49_213520 | 138 S | TIGMSEEEA | 0.995 | unsp |