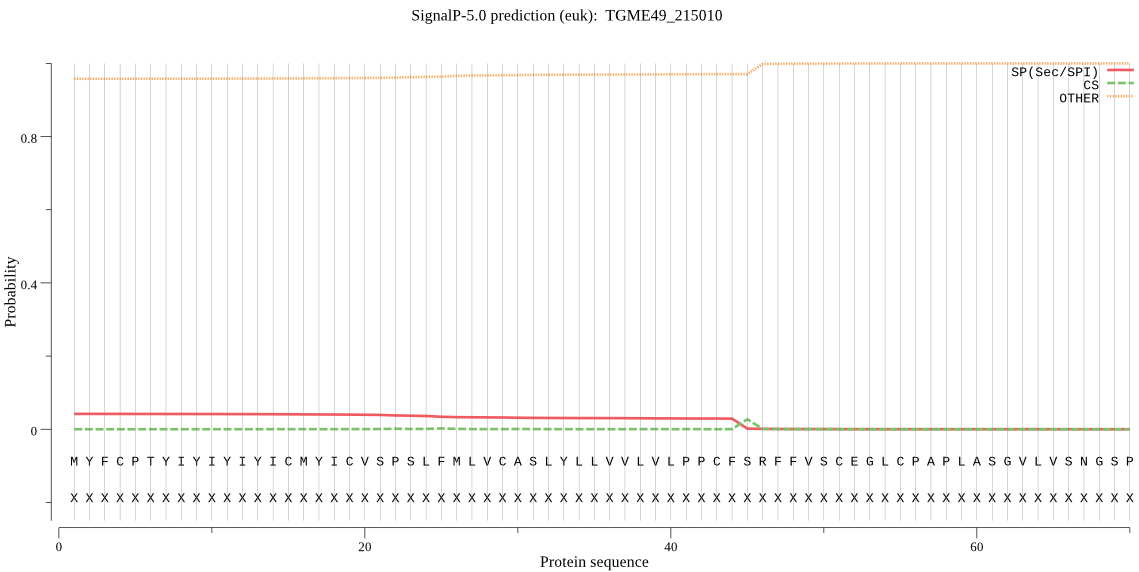
Fasta :-
MYFCPTYIYIYIYICMYICVSPSLFMLVCASLYLLVVLVLPPCFSRFFVSCEGLCPAPLA
SGVLVSNGSPGASLDFIFCWTADRLLCSVSSSSSISSFFSDRSSQTGNLCLRSFLLPRLL
VSAVLFSPRFSNSASLLSLTSFSTMALTAVSSASSRSPPSSPSANPHASPALVASALPSS
RGTTHRVSPPIPTAPSPPLSRPVYLPPQYLVQNAPQTCHAVPVPPVHAVPVPPVHAVPVP
PAHAVPVPPVDSPSNGEAREGQELHGCLCCLRSFSPTLFVSAVLGTWMFAGGFGGEIVLA
LATLPAIFIVYGLHSSVKARSLSGVSLLEFFWLGAFVSVFVAMCLELLANATLYNALLSC
LPPLPSPSRASQWALNDLRSLASGSADISIAPSSDSPLFHPPPADPLSPSAPSSSLFLSL
LQFLPSWTFAPLLAPLSLPFYFPLFESKAAPGCLRHPSLQSDSSAALHSSSSSSVLSASA
FSASSLFSASPLSAFSFLPDHSGERWQRLLSTLNGRHAHGALPTLGCSLSLVAFMIFCVG
FVEEFAKLVVLQRLQVLPVPLPLAWGGAEEASSERRNALEQRTLGGCCRPFWTRYVKHPV
GVCLAGCAAGAGFAVAENLSYTLGRRGIFAEHLIVAIVRTFTAVPSHIANTGMAAANLAL
AWHAACALPAASPFFSSHLEAHQQPLIPGSSRASGHALPSFWRCLLPSLLVPSLLHGTYD
AALRLSAAYAQPGPAVDDNAAVVAAVFLLVSFLAWLLTLLLFWQKWRRVCDLPAFGGSCA
AAFGPTAPELRRVHPQASGWDSRAQWVSPQAAQAPETGGDDLLAGSAAQSAPLFCSPSPQ
RFAPYAVAPAFPFATLAAGTGETGVRADVLVWEERRREANRRIQQHANSLHATAPAQFSP
APAAVPRAEKPTGEWASLAQR