

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
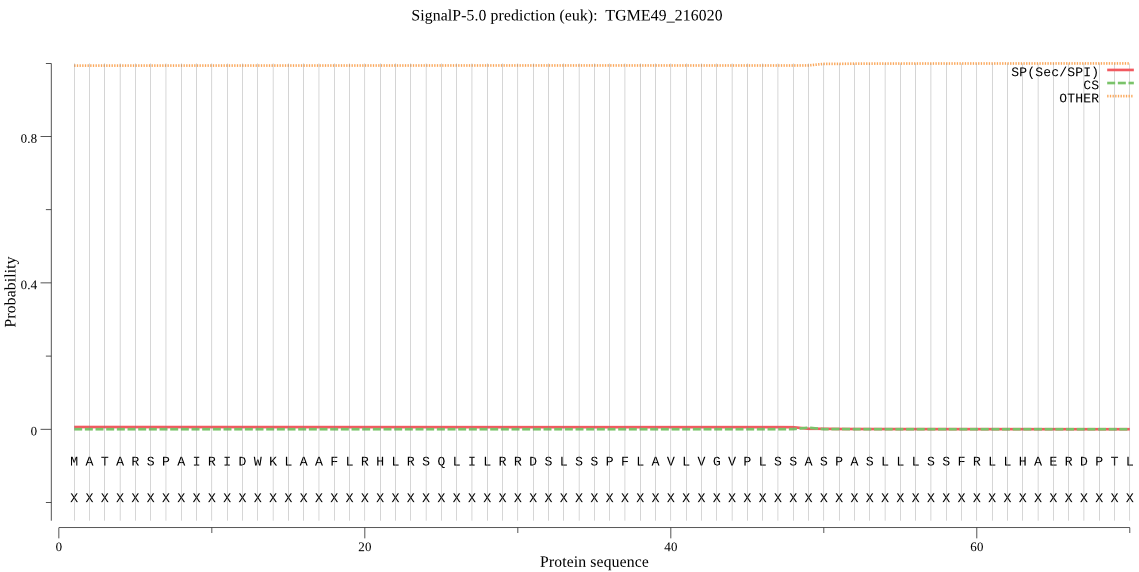
| TGME49_216020 | OTHER | 0.882782 | 0.033862 | 0.083356 |
MATARSPAIRIDWKLAAFLRHLRSQLILRRDSLSSPFLAVLVGVPLSSASPASLLLSSFR LLHAERDPTLAQLFTASTPSVSSSLREALRRDLATITFILPDGLRILGLLAFSPADGPAV SAVYPSVSAVAEAISTQLAFLAQKELSSLSEDAGVAASHRLSFVCSLSSDAEKTTGASVF PWTFVSLLPSLSSSSPSSSSSPSTVEESAGARLTVSVEDGSSLLRSERFLFLRSRLALSL RLETPFLVTLSRGLGDARHGATDAVSSLEAERELAAGEDRADFLPCLPSTTATELLAALR EQEAALAAPLGLFFCMPNGVYVHPTDGVEAASARLEKSPSRLGGCDTSVWTAVAPAASKA KHVVVGEAKIADEDAERNNTEGRLQTAFSTHAVLDLPWLQQASQGATQDGRLPLASLSAI SIPAFLSSLSSASESPHGWAYTPECGAAQVSLRYIQVFLDSCILVPGTASLASCANRFLR AFRRQLRHAALQILHSSSSSSSSSSCSVQNILTTSLTAALMSSGDFAASSASAASACAPS PPLFPRVLCFDASRCRVPREEQPASLSFAFHSVKLPWCLHPLLSWNNEAQVARRHWLSLL GVLKDLPAFRLSAALPWNETQAERGRGAHKLVNPHRSLVTGPKWLPQTRAVTCLVQGDFA YYHYCQDNERDEGWGCCYRSLQLVISWFQRQHFTTKPVPSIADIQRLLKKFDLAHENLEI GSKAWIGTVEGSYVLNWYLHVPSKLLHLTSASDLPSHAATLKEHFDSVGSPVMMGVGDYA YTMVGISVDSETGEAAFLIVDPHYAGDDGDIDKILDKNWIGWKKTNFFEKTAGTKFINLA LPQICTEGGDLFV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_216020 | 113 S | LLAFSPADG | 0.995 | unsp | TGME49_216020 | 113 S | LLAFSPADG | 0.995 | unsp | TGME49_216020 | 113 S | LLAFSPADG | 0.995 | unsp | TGME49_216020 | 195 S | LSSSSPSSS | 0.994 | unsp | TGME49_216020 | 197 S | SSSPSSSSS | 0.991 | unsp | TGME49_216020 | 198 S | SSPSSSSSP | 0.991 | unsp | TGME49_216020 | 203 S | SSSPSTVEE | 0.995 | unsp | TGME49_216020 | 216 S | RLTVSVEDG | 0.997 | unsp | TGME49_216020 | 267 S | DAVSSLEAE | 0.99 | unsp | TGME49_216020 | 501 S | SSSSSSSSS | 0.991 | unsp | TGME49_216020 | 503 S | SSSSSSSCS | 0.991 | unsp | TGME49_216020 | 32 S | LRRDSLSSP | 0.997 | unsp | TGME49_216020 | 84 S | SVSSSLREA | 0.995 | unsp |