

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_216155 | SP | 0.264741 | 0.734756 | 0.000503 | CS pos: 21-22. VSG-TS. Pr: 0.7253 |
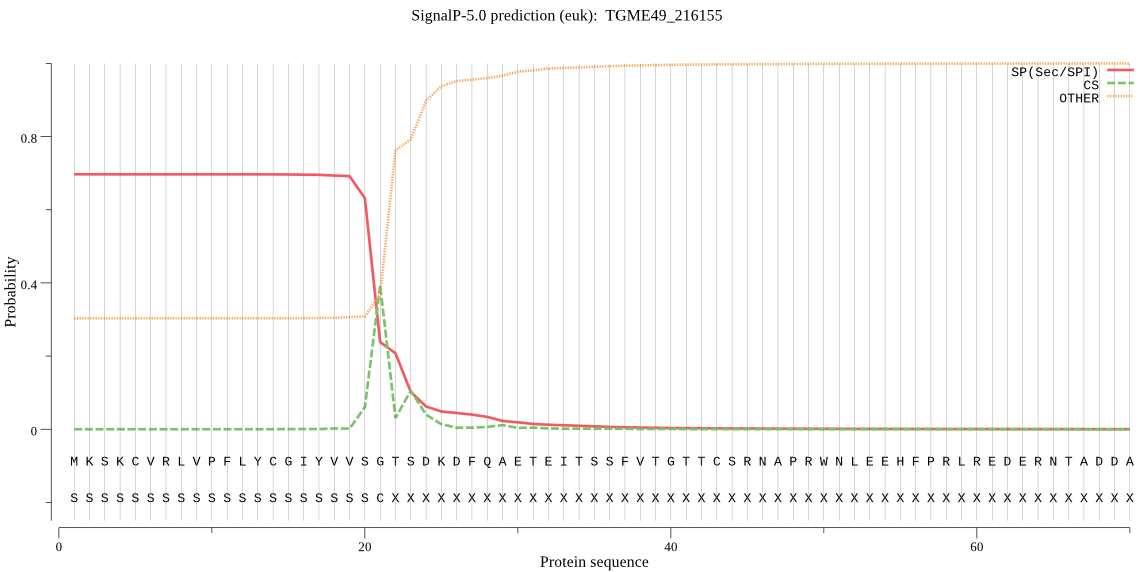
MKSKCVRLVPFLYCGIYVVSGTSDKDFQAETEITSSFVTGTTCSRNAPRWNLEEHFPRLR EDERNTADDAVLDRLEERAIAFAEQHRGTLDRSLGDALEEYEKIDEELDRFALYWSLKTS QNLADDTLRRKRSKADTRASR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_216155 | 133 S | RRKRSKADT | 0.997 | unsp | TGME49_216155 | 133 S | RRKRSKADT | 0.997 | unsp | TGME49_216155 | 133 S | RRKRSKADT | 0.997 | unsp | TGME49_216155 | 23 S | VSGTSDKDF | 0.996 | unsp | TGME49_216155 | 93 S | TLDRSLGDA | 0.991 | unsp |