

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_218540 | SP | 0.011193 | 0.988308 | 0.000499 | CS pos: 21-22. ALA-IL. Pr: 0.2368 |
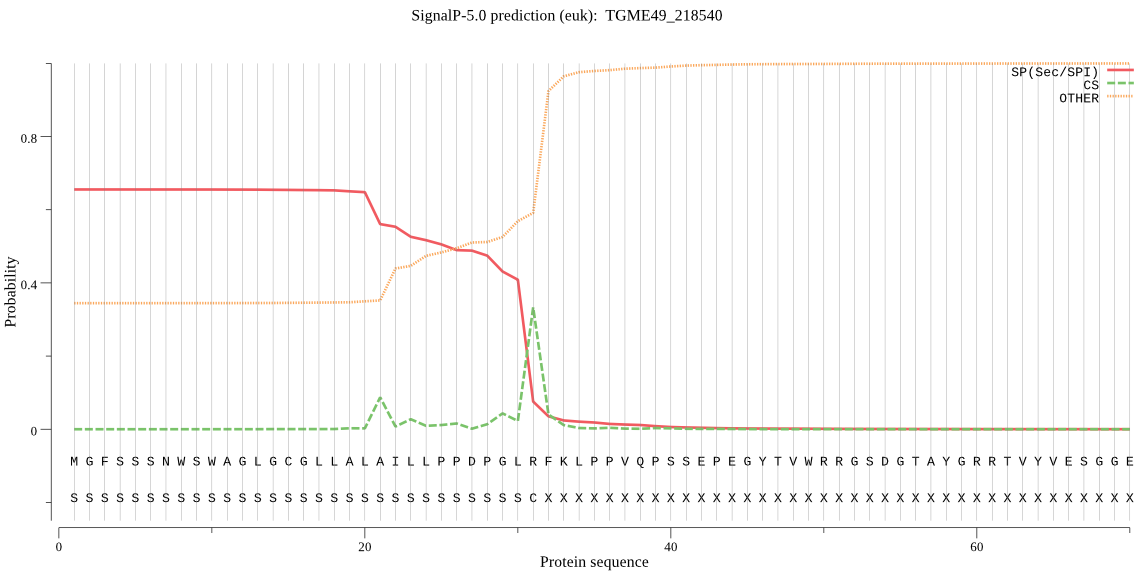
MGFSSSNWSWAGLGCGLLALAILLPPDPGLRFKLPPVQPSSEPEGYTVWRRGSDGTAYGR RTVYVESGGEKLHGWLYLPPKPRQNVPIYVVCHGFGAVMPVADVAFAEKLQEYGMAAIAF DYRTWGFSGGAPRQVVDPHMQLQDIRAVLQHIVDTGGFQGTVDAANIHLFGTSYAGGHVL VTASQLASEKSPFLRRVRSVTSVVPLIDGQAQTKKALQQRSFFRTLRYAAAILADLLRHA VGSRLQPVYLHVAGPRGASTLSALELQGGELEIWSNRVLDGSRTVAWTNALAARSIFMTS QYRPISYLENIQTPTLLVEAQHDDTCIPELTEEALKIINSRGANVREHRGSKSTEKGRLA ELYRMPVNHFEIYLSPHLQDLINTTVAFAKRHGASDEASVDKFAGGDSPKKS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_218540 | 408 S | AGGDSPKKS | 0.994 | unsp | TGME49_218540 | 408 S | AGGDSPKKS | 0.994 | unsp | TGME49_218540 | 408 S | AGGDSPKKS | 0.994 | unsp | TGME49_218540 | 351 S | EHRGSKSTE | 0.995 | unsp | TGME49_218540 | 395 S | RHGASDEAS | 0.991 | unsp |