

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
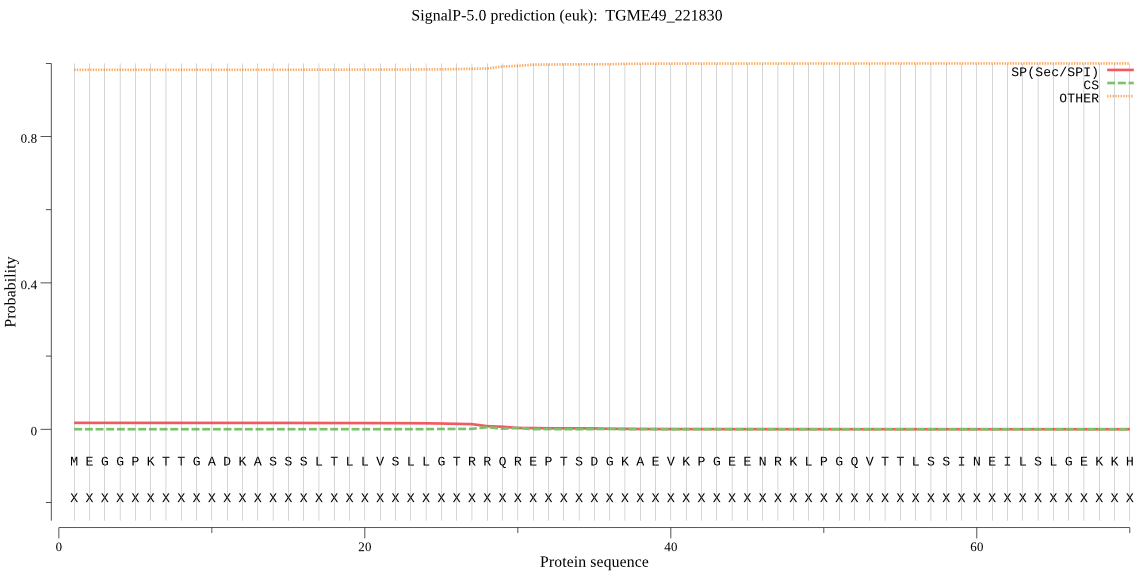
| TGME49_221830 | OTHER | 0.998205 | 0.001628 | 0.000167 |
MEGGPKTTGADKASSSLTLLVSLLGTRRQREPTSDGKAEVKPGEENRKLPGQVTTLSSIN EILSLGEKKHLELSIRNPYPYDGQLIVTVDTGEVSPPAQAQTASFTHHGGFGHADFTWED LAALVHRHKEEGKKQTLEESKKNEKETDRPNAGEEEESRDVQDSPFVVRPSSQANRRQQE REDQRPLSTSGASNSYSPFRPFSSSSFSRVNSAYSSESLSFSPWSSEDSSSSDSSYSAFP SSRRRVAEKKSITEVAEEVERNEEAVEDDDIRQDAGDTEGEEGAAQSEEEQEPETSKTHE EESLCTEEGYSTETEKSAPGRRTQQRSSATTNSGDETDWDVRERAGGKPSGEFGTSSPSK HKRPPISVARTDLPPRLVIKLKEGTRALEFERRTREFQRQKWVDQGFVEPNTYGDTSVAA GVSVTVDDPYEIQKHLRGFFSFNEQQAELVSSLSHLKQLDLLVLDIEESRRLYSSDTTKL KDAFSQVSDVEFVEFDEVVYALEIIPNDQLFSLQWNLFYPRNRMVDVNAPQAWEVVDEVL REKREREGLRREDEDWKGRVVAVIDTGVNYLHPDLKNQMWVNVKELYGVPGVDDDGNGYV DDVYGYDFVNDHGDPMDRDGHGTHCAGIIGAQANNGLGIAGINSRTRIMALKFMEGETGY VSDAARSLDYAIQNGASISSNSWGNAARLPRTMIEAVHRSAEKNMLFVTASGNAGGIDST SENRDENRFGRDLDEEPLYPAALANSNVLTVGAVGQSGELASFSHFGKKTVDLAAPGMNI FSTWLLDSYEYKDGTSQACPLVAGVASLLWTMKPEASVEEIRRILIVSSKRLPTIQNTIA HGLVDAYAAVVTLQETLGQWITVHAPSTLSPRSTATVSLDIRGKQNGFYRSQLRLLANYQ GTPLNLAIVPIRLQVAESPILEFPPVIVLDPVLRNVNSLPVALPVRVPTDQGAVLVKVLK IDATAADTFFHLSPPGGEVRIENGRFLSPIKVGCRMLSPGTEDGILTLRARLADASPSSS PSSSSSSSSSSPSSSEPVERLYTVRLRCVAIDVGVAPVFLDLTVPPDSEVMQEFSIEKGE RLLQREVRYSVEFVSVVEQSEEGKKNFRGARETGVERLRGSDSKFDSGGSAPTRNSGGNM SDWAWNRESERIPGQNRSGEEDSNEALGDAGYAYLINGADPPPGLPPVKYQWNDISASGQ PLFASGRLSRIRRVTLAFDFPFWGSPFRQLFVSPTGFVSVENVQGPELWKPPSMPDKEAP NGVLAPLWMDFPPSDSPGSNIFFQTWQDRAIVQWENVHVRKRSFQRSGVPTAGATFQLVL HASGRILFYYKSVAPTYTPRDWAAAVMGVESPDGLRATKISEGVPQSEMAVLLFPSSFAS SSRLSSRAKSSSSSRGLSFGGSASSAGRNLQLQHESGVMDVSGTKTRVSFTVNSKNFKAG ETVSGFFLVTAMFLDSSLNKVSGSGAVLVSIKVEAPKASSGDSAGQEQQTKAKTAQCPLS EELAAEQWNELLDVFVRLPQATLVGAEEIAKNSPLSLGRCALLCLEDPSCARFSFNWIKT ECVRYSRNPEEKSDATALQRSPRQNAAERGQRTPAVQTVVDSWTDLFERREEANDEPLGP PPFLPYGRMPPPFPPGSPGPYGPPPGSPVPYGPPPGSPGPHGPPPGFPGFLPLPSAGPLD APPKGTPENPPLPSASAPSDANESTPISVTSSSSPSPVSSPTPSFSSPPSPASSSSPASS SASSPPSSSGGGGGSEAGRQAQNAGEKEDAKSEHEEQKPQPRPPPRPAPVPRPPPSRSPF PPQLAQRRPPMPPPMGGGRPPRPGGGGQQGLAGPYAFLPPPPASLPPPSPSSPSSPSSPS SPPSSPSSPTSSPSSQAPPPPSPPYPAPPYGFPPSPPFAGGGYPPPYLPPYPLYPPSPYF MPPPGPGFGPSPPQQAQAPPNAGVA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_221830 | 225 S | FSPWSSEDS | 0.996 | unsp | TGME49_221830 | 225 S | FSPWSSEDS | 0.996 | unsp | TGME49_221830 | 225 S | FSPWSSEDS | 0.996 | unsp | TGME49_221830 | 230 S | SEDSSSSDS | 0.997 | unsp | TGME49_221830 | 234 S | SSSDSSYSA | 0.993 | unsp | TGME49_221830 | 287 S | GAAQSEEEQ | 0.996 | unsp | TGME49_221830 | 311 S | EEGYSTETE | 0.991 | unsp | TGME49_221830 | 328 S | QQRSSATTN | 0.992 | unsp | TGME49_221830 | 817 S | KPEASVEEI | 0.997 | unsp | TGME49_221830 | 870 S | PSTLSPRST | 0.993 | unsp | TGME49_221830 | 1016 S | LADASPSSS | 0.991 | unsp | TGME49_221830 | 1020 S | SPSSSPSSS | 0.993 | unsp | TGME49_221830 | 1023 S | SSPSSSSSS | 0.992 | unsp | TGME49_221830 | 1027 S | SSSSSSSSS | 0.993 | unsp | TGME49_221830 | 1031 S | SSSSSPSSS | 0.997 | unsp | TGME49_221830 | 1033 S | SSSPSSSEP | 0.996 | unsp | TGME49_221830 | 1034 S | SSPSSSEPV | 0.994 | unsp | TGME49_221830 | 1068 S | VPPDSEVMQ | 0.993 | unsp | TGME49_221830 | 1090 S | EVRYSVEFV | 0.994 | unsp | TGME49_221830 | 1158 S | GQNRSGEED | 0.99 | unsp | TGME49_221830 | 1209 S | SGRLSRIRR | 0.993 | unsp | TGME49_221830 | 1385 S | SSRLSSRAK | 0.996 | unsp | TGME49_221830 | 1390 S | SRAKSSSSS | 0.997 | unsp | TGME49_221830 | 1392 S | AKSSSSSRG | 0.991 | unsp | TGME49_221830 | 1429 S | KTRVSFTVN | 0.991 | unsp | TGME49_221830 | 1581 S | ALQRSPRQN | 0.993 | unsp | TGME49_221830 | 1602 S | TVVDSWTDL | 0.994 | unsp | TGME49_221830 | 1716 S | SSSPSPVSS | 0.995 | unsp | TGME49_221830 | 1744 S | SSASSPPSS | 0.994 | unsp | TGME49_221830 | 1772 S | EDAKSEHEE | 0.996 | unsp | TGME49_221830 | 1852 S | PSPSSPSSP | 0.993 | unsp | TGME49_221830 | 1855 S | SSPSSPSSP | 0.996 | unsp | TGME49_221830 | 188 S | QRPLSTSGA | 0.996 | unsp | TGME49_221830 | 1858 S | SSPSSPSSP | 0.997 | unsp | TGME49_221830 | 1861 S | SSPSSPPSS | 0.995 | unsp | TGME49_221830 | 1865 S | SPPSSPSSP | 0.995 | unsp | TGME49_221830 | 1868 S | SSPSSPTSS | 0.997 | unsp | TGME49_221830 | 1872 S | SPTSSPSSQ | 0.991 | unsp | TGME49_221830 | 212 S | SRVNSAYSS | 0.991 | unsp |