

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_223510 | OTHER | 0.999062 | 0.000935 | 0.000003 |
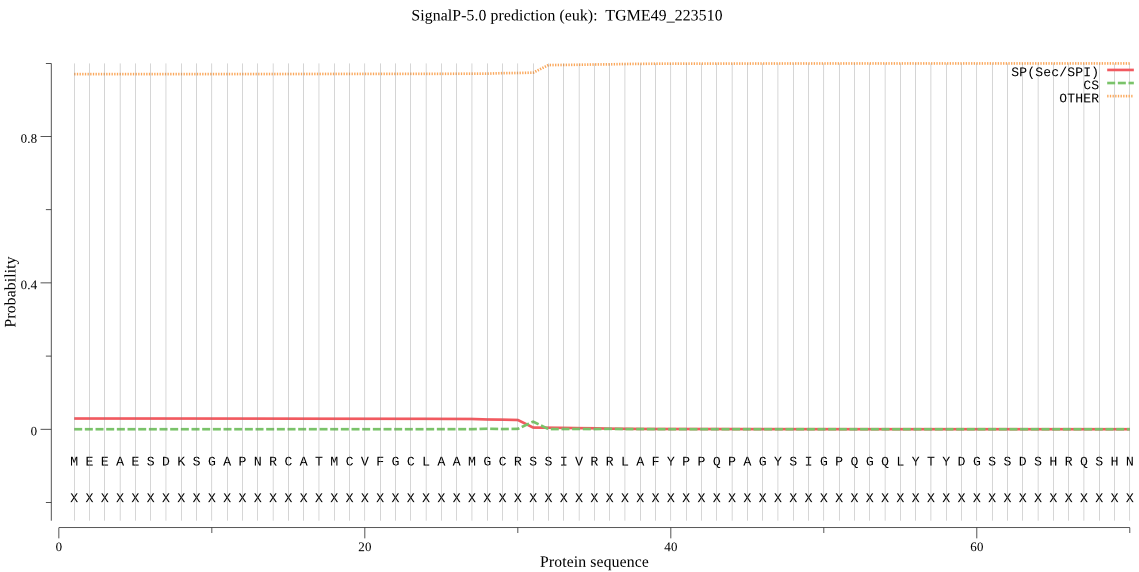
MEEAESDKSGAPNRCATMCVFGCLAAMGCRSSIVRRLAFYPPQPAGYSIGPQGQLYTYDG SSDSHRQSHNQSAPPADSINSASGAATAQPDNPPAQQLQRESMQQLLQRTGLPERLKVLS IPCGRVKLAALFIYHPLSPSSSSTGPLSESESGQGPASSVLPAGEGNAREQTRGAAGPLA GGADSCDDQVMHESAKRLPCIIFSHGNSTDIGFMFGLYYRLAYKCRVNVLAYDYSGYGCS GGKTSEKALYRNIRAVWTYATQMLHVPPRQIILYGHSVGSAPCCDLAMREKSFPVGGVVL HSSIASGLRLFFDDIKKSPWFDAFPNVEKLKKVKRTPVLIIHGQLDRQVSWIHSQRLQDA AAAADADWFKELERKRAREKASGKKRLTESPNEETQLNAAAMDELHSQRVQVWWVGNADH NDVEQKSGEAYYKHIGDFIHFCNVWASNCHGG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_223510 | 148 S | TGPLSESES | 0.995 | unsp | TGME49_223510 | 148 S | TGPLSESES | 0.995 | unsp | TGME49_223510 | 148 S | TGPLSESES | 0.995 | unsp | TGME49_223510 | 6 S | EEAESDKSG | 0.996 | unsp | TGME49_223510 | 138 S | YHPLSPSSS | 0.995 | unsp |