

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
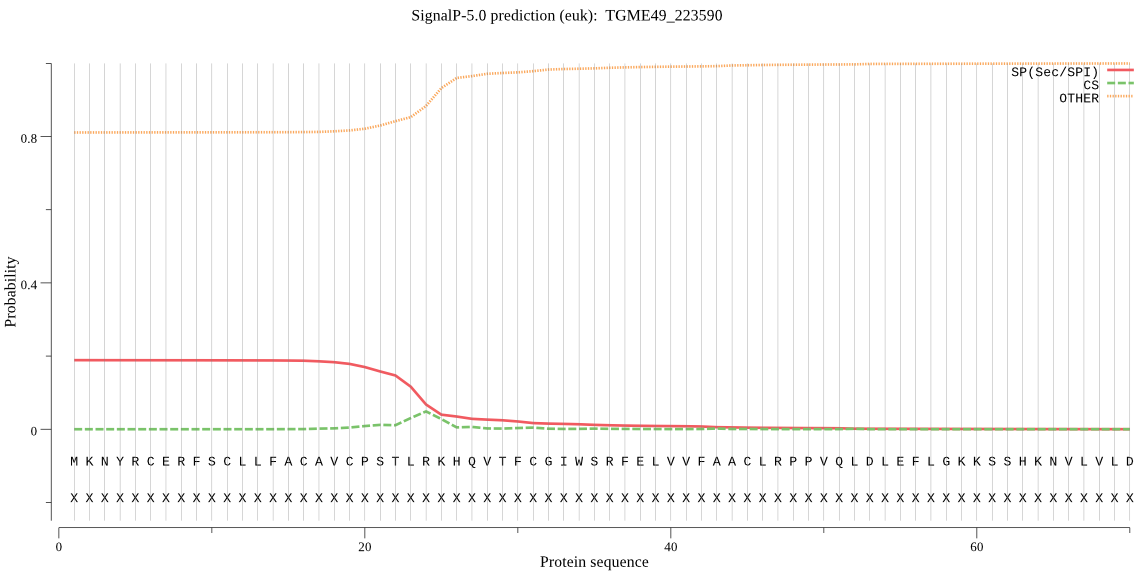
| TGME49_223590 | OTHER | 0.598972 | 0.392622 | 0.008406 |
MKNYRCERFSCLLFACAVCPSTLRKHQVTFCGIWSRFELVVFAACLRPPVQLDLEFLGKK SSHKNVLVLDGEQFLFGVHESRTQSRLPSTGRCAKPLFPTPPRGLSRTFSTRCLCPPIRH DVATMASSSCVAFSYDDLGLPVVAASSPAGLPVPSLSASGRGPTAVSTGTTIVAVSFKGG VVLGADTRTSAGSYVVNRAARKISRVHERICVCRSGSAADTQAVTQIVKLYIQQYAQELP KGEEPRVEAAANVFQSLCYQHKDALTAGLIVAGFDKVKGGQIYALPLGGALVPMQYTAGG SGSAFISAYMDANFKQNMTQEEAVNLVKNSVAYAISRDGSSGGMVRVVVITEDAMLEECV EGNKLPVAP
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| TGME49_223590 | 176 S | IVAVSFKGG | 0.994 | unsp |