

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
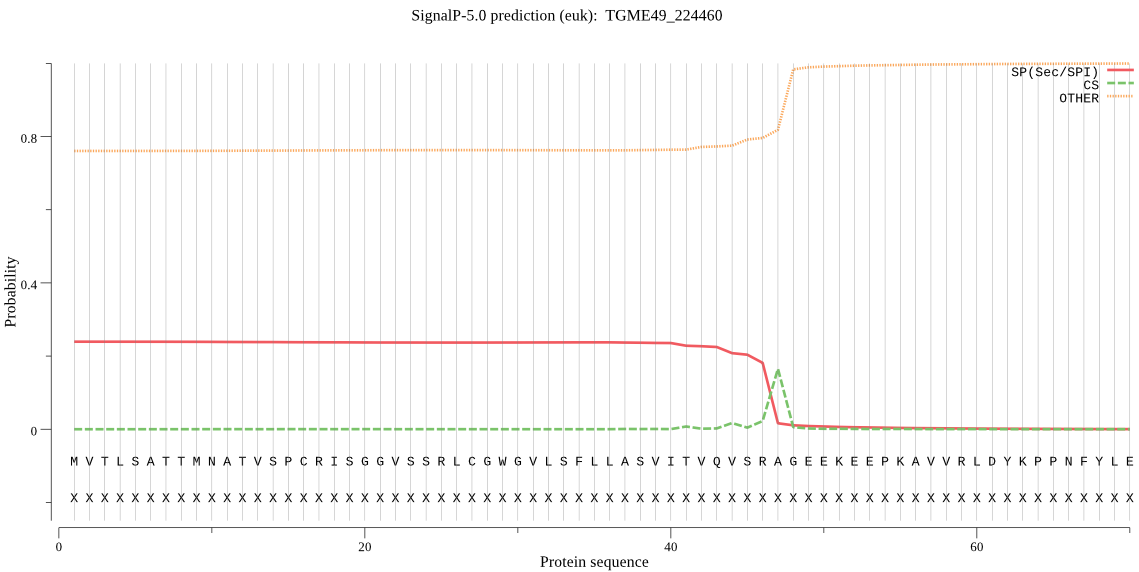
| TGME49_224460 | SP | 0.376312 | 0.446582 | 0.177106 | CS pos: 47-48. SRA-GE. Pr: 0.6552 |
MVTLSATTMNATVSPCRISGGVSSRLCGWGVLSFLLASVITVQVSRAGEEKEEPKAVVRL DYKPPNFYLEDIVLDFNLDDEDTTVEALLTLYRRAGTEPMNLVLDGDESVILKSISLATT SSPSLRGQGRVTFTELDGQMPTDSQFQFRIVDGNLMVSKFLLPREAEVRFYIRTLVSINP KANLALFGLYKAGDIFVTLNEPSGFRRITYGVDRPDVLATYTVTVTAPRHLPILLSNGDK VLSGYAGENRHFAKFVDPFPKPSYLFALVAGDFASVSGEFVTMSGRSVTVTIYAQHHQRN QLQWALRSLLRAMRWDEETFGREYQYSEFRVLCVEVFNPGAMENTSLNIFTCSLLLADPK LTTDADHRLIVDVVSHEYFHNWTGNRVTVQDWFQLTLKEGLTMFRNNSFTEETTSRAMKR IGDVADILSIQFREDSGPFAHPIRPETYKSIENLYTRTVYLKGAEVNRMYRTMLGPKGFR RGMDLYFKRHDGKAVTCDDLRAAMADANDKDLSQFERWYSQAGTPHVTVSSFIYDAAERK MHLTLKQHTPPTPGQDKKLPLQIPVAVGCIGKTSKRDVLSPPTQILELTEEEQAFVLVDV GEDCVLSVLRDFSAPVKLIQPLQTDEDLAFLVTYDSDNVNRWQAAQTLSRKVLLTRISEY QQRVEEIEEGRVSESDLFSKLPTQYVQTVRDIVIAPESSMGKDIKSLLLSLPTKAQLELA VDSIDPDAINAALASVRRDIVDALGEEMLQLYTELTLPAGTEESGADIEHWGRRALRNEL LRFLTASFDQKSAKLASAHFDRAMVMSDKVAALTVLTEIPGQERDEAFERFRQEVAGNAV LMEKWMSLQAMSAAADTLERVAQLRQHSAFHPFIPGSAYSLFGTFSSSSPQFHSKTGAGY ALVLDFLTMIDEHNSLVSSRIAGVAFANWKRFTSPRREMLKEVLKKLLKKEKLSVGLKEV LEKALAGEDD
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_224460 | 934 S | KRFTSPRRE | 0.994 | unsp | TGME49_224460 | 934 S | KRFTSPRRE | 0.994 | unsp | TGME49_224460 | 934 S | KRFTSPRRE | 0.994 | unsp | TGME49_224460 | 124 S | TSSPSLRGQ | 0.993 | unsp | TGME49_224460 | 673 S | EGRVSESDL | 0.998 | unsp |