

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_225580 | OTHER | 0.999983 | 0.000010 | 0.000007 |
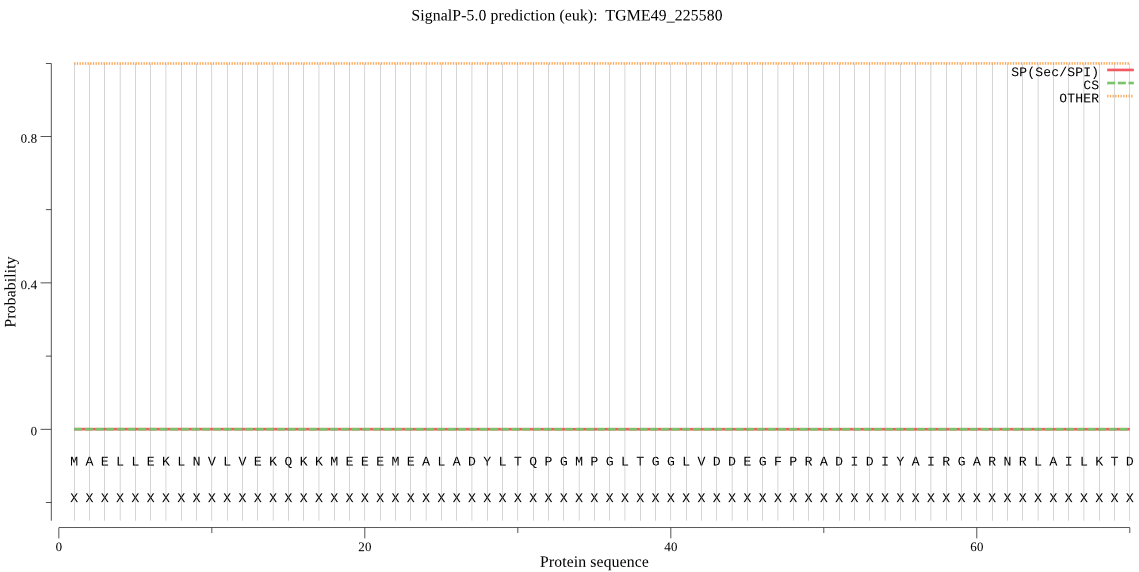
MAELLEKLNVLVEKQKKMEEEMEALADYLTQPGMPGLTGGLVDDEGFPRADIDIYAIRGA RNRLAILKTDYKEVRSQIEKELFALHAQGPVAVPRTGCVSTGDALNAASLSPDSNTALEE NPFIPFAKISELHENSPASKAGLRLDDLILQFGAVFIRRKSLPEPSEGDKQQVAADPGGD RPGCSSVEQVFERLPHEVGNHTGEEIDITVFRNNAILNLKLIPQTWEGMGLVGCRFTPVT KGML
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_225580 | 100 S | TGCVSTGDA | 0.993 | unsp | TGME49_225580 | 161 S | IRRKSLPEP | 0.998 | unsp |