

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_226420 | SP | 0.428227 | 0.530194 | 0.041579 | CS pos: 49-50. CSG-EI. Pr: 0.5410 |
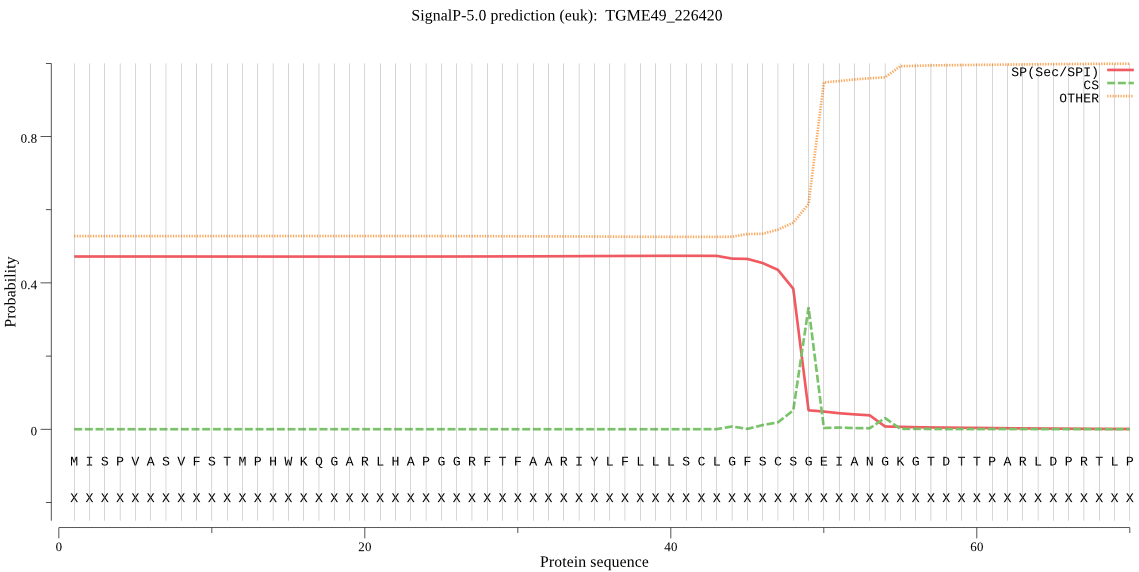
MISPVASVFSTMPHWKQGARLHAPGGRFTFAARIYLFLLLSCLGFSCSGEIANGKGTDTT PARLDPRTLPRYNLKRHFPYSSIFDPAIDRDIEALSRRAREFKERFKGTLETTLLEALIK HEQLDEAVDFVDYYIALVAAVNTTNEEIRKRVNQLESAIAADVQPYRLFVNLELGAMPEA ALQKQMAANGNLSFYSGFLGAARRRAPYVLSEDVERALLVRKPMTVQVATEYYFKQLGDA TFDMNGEKLILSMILAFIMDNSQEKRWAAQKAVKQGLERHKITNFAALSLNVVAGSWHME RKERGYERLRSSRNVANNVSDKTIDALIHAGETVAVSLTKRYFKLKKKILKNRAGVEVFD FADRLAPLPLKSSDRLYDWKESTRIVRDAYKSFSPTMALMFDKLLEEERIDAPAVPGKRT PACYLGTPTTGPFIILEHVGQPRCVETLAHEAGHAIHSMLSYEQGNLQLAPPLTIAEMAS LMGERIVFNDLLRRTTDPEERLSHLLRYLDGWTSTVTRQLMFDDFERRVHDARAQGTIPD GAFTTMWKESLEKYFGAEGEVFDSYANSEADWARIPHFHNTPFYVYAYAFSDLAVGSLYR VYKAHPEGFEQKYLDILRSGNKKSFEEIMKPFGLNPSAETFWADAVEATGGTVIDEAESL ALKLGFV