

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_228190 | OTHER | 0.910150 | 0.002342 | 0.087508 |
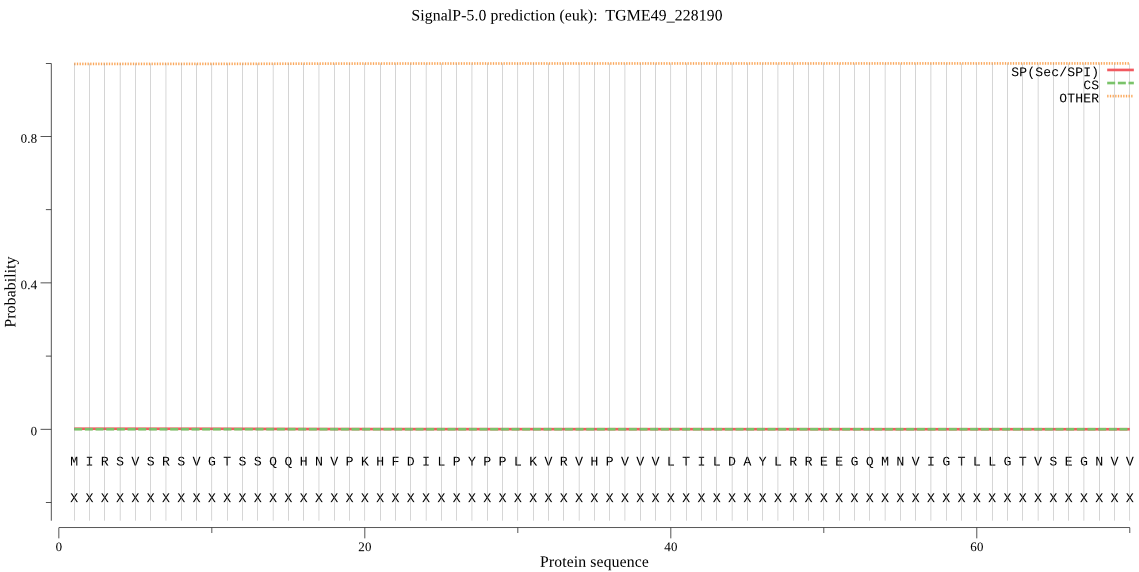
MIRSVSRSVGTSSQQHNVPKHFDILPYPPLKVRVHPVVVLTILDAYLRREEGQMNVIGTL LGTVSEGNVVDISDCFVDRHSLTDEGLLQIIKDHHETMYELKQQVSGSGTKDIVVGWFCT GSEMTELTCAVHGWFKQFNSVSKFHPQPPLTEPIHLMVNTTMDRDNLSIKAYMQVPMNMA KDACFQFQELPLELFASSSDRAGLSLLLKVREANRRHRQQHDGSRASALSSSSTSSAAAV AAISDTPGVAGVPVIKQGLGAALEKLSDQLNKCGAYVRSVLDGSEKADPEIGRFLSKALC VEAVQDLEVFEQMCQNALQDNLMVVHLTSLARLQFAVAEKLNTSFF
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| TGME49_228190 | 81 S | VDRHSLTDE | 0.998 | unsp |