

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_229710 | OTHER | 0.868583 | 0.124494 | 0.006923 |
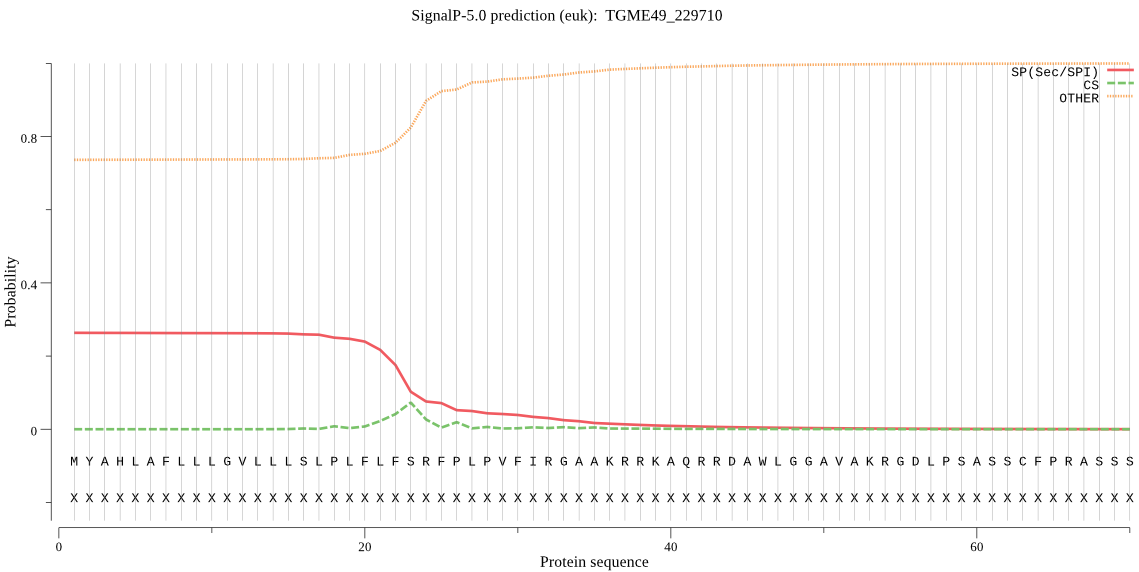
MYAHLAFLLLGVLLLSLPLFLFSRFPLPVFIRGAAKRRKAQRRDAWLGGAVAKRGDLPSA SSCFPRASSSPLSVSPLSFCLSHLLSLLLPFLEISFLAPALRATVLYWLSVFPAFSSLRF FPAPSTLRSLLPCSFGSSLLRFLMGKKKNGRGRGTARRPGKERRGDSDGEQPEKSDRREK KEKASRDTAGCSSFSSQLQAYGLELELIVPDGNCLFRAFADQHCGKQERHGEYREKAVDY IEAHAEDFQCFLSEEEESFKKYVNRMRRLGTWGSQVELQALSQVYEVSLFVHVANGVEPV GRWHGGAGAPQGDTCGKKKGRGVGKKGGAKPNQRRGRGDDEGDGEDWNICKMENFSESHP CLQLAFHVNHEHYNSIRVRGQTPGKMLTLAQVRRVLNMKAESDRESGETGEETADEAAPE PVEEEEEEEEEEEEAATEKRECVERGTEEDLEEADKSVEGERGESATRRVESDTQVRGPA ESVRAGQREQEGEKDEGVFPGVQPATEASGNCVRNLLESKGEEHEESREQVEDAATQSGD GSAKSREETVELTLVEAGKSEETEGTASEEEKCGDARKQETSAAEETAEQIPEGDVKRHG ARGKRRIQQRERGEQSTTEDRKTADRASKNDALREKGEASSFAPDKHAQEDARSQHTRSS SSASNGSSSSVSVSSSSALSSTSSCCLSLPQSSSPSSSSSRSSFSSSSVACCSFSPASVP VSCSSVAASASVRPLARSVSASPRWATVPRRSRSTGALVRSSGEVEREASFCFASRQCCL VPSSGNASVSDSPASSDEAFSAPAHAEAFSLHGGLPASGSPSRENSSSEDGASRFLHSHR LSECRGDHRRPCAGFHADLREGEKDGFQNRRVKRRNSLRVCSTSSWSPSRSLLSGRSSRI CATCGYRMQPLPSSLKQSARPSPLSESPSLPSSVLSGASCSAAAVASSPSALVPSSRCSS EVLPNGDEETGEEKDEREARPSPLVCV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_229710 | 175 S | QPEKSDRRE | 0.994 | unsp | TGME49_229710 | 175 S | QPEKSDRRE | 0.994 | unsp | TGME49_229710 | 175 S | QPEKSDRRE | 0.994 | unsp | TGME49_229710 | 402 S | MKAESDRES | 0.995 | unsp | TGME49_229710 | 406 S | SDRESGETG | 0.998 | unsp | TGME49_229710 | 457 S | EADKSVEGE | 0.995 | unsp | TGME49_229710 | 519 S | NLLESKGEE | 0.991 | unsp | TGME49_229710 | 545 S | GSAKSREET | 0.998 | unsp | TGME49_229710 | 568 S | EGTASEEEK | 0.997 | unsp | TGME49_229710 | 628 S | ADRASKNDA | 0.998 | unsp | TGME49_229710 | 662 S | RSSSSASNG | 0.994 | unsp | TGME49_229710 | 672 S | SSSVSVSSS | 0.992 | unsp | TGME49_229710 | 698 S | SPSSSSSRS | 0.99 | unsp | TGME49_229710 | 699 S | PSSSSSRSS | 0.991 | unsp | TGME49_229710 | 703 S | SSRSSFSSS | 0.996 | unsp | TGME49_229710 | 742 S | SVSASPRWA | 0.997 | unsp | TGME49_229710 | 752 S | VPRRSRSTG | 0.995 | unsp | TGME49_229710 | 754 S | RRSRSTGAL | 0.992 | unsp | TGME49_229710 | 762 S | LVRSSGEVE | 0.996 | unsp | TGME49_229710 | 788 S | SGNASVSDS | 0.997 | unsp | TGME49_229710 | 792 S | SVSDSPASS | 0.993 | unsp | TGME49_229710 | 795 S | DSPASSDEA | 0.996 | unsp | TGME49_229710 | 822 S | SGSPSRENS | 0.993 | unsp | TGME49_229710 | 826 S | SRENSSSED | 0.995 | unsp | TGME49_229710 | 827 S | RENSSSEDG | 0.998 | unsp | TGME49_229710 | 842 S | SHRLSECRG | 0.995 | unsp | TGME49_229710 | 877 S | KRRNSLRVC | 0.991 | unsp | TGME49_229710 | 887 S | TSSWSPSRS | 0.994 | unsp | TGME49_229710 | 894 S | RSLLSGRSS | 0.992 | unsp | TGME49_229710 | 959 S | SSRCSSEVL | 0.992 | unsp | TGME49_229710 | 155 T | RGRGTARRP | 0.99 | unsp | TGME49_229710 | 167 S | RRGDSDGEQ | 0.998 | unsp |