

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_231900 | OTHER | 0.998743 | 0.000138 | 0.001118 |
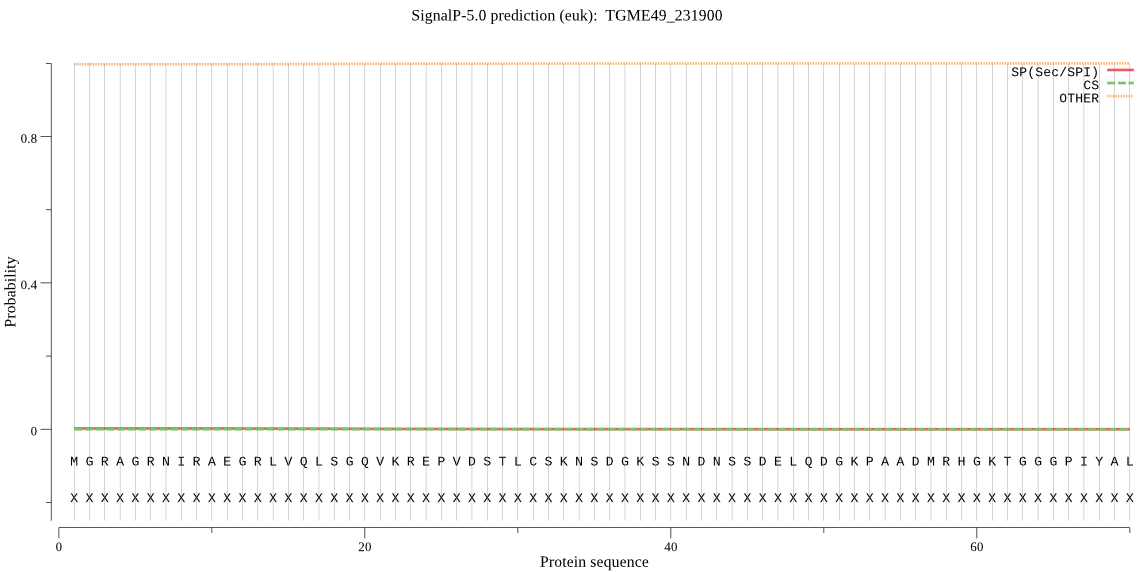
MGRAGRNIRAEGRLVQLSGQVKREPVDSTLCSKNSDGKSSNDNSSDELQDGKPAADMRHG KTGGGPIYALDFMHFDSLLTPRERTLRGRVQNIVSRYVEHAVLPLYEAAAFDSELVEICK KFGPAGLQIKGYGCPGLTHTEAILMAVEMARVDASFSTFYLVHCGLAMQSIAVAGTQEQK AMWLPKMSRLECIGSFGLTEPNHGSDATGLRTSAKKVEGGWILNGQKRWIGNATFSDIVV VWARDVHSEKILGFIVEKDFPGFSATKIENKVSLRIVQNADIVLKDCFVPDSHRLQHEGF SGATAQVLESSRAVVAAECVGVLLGAYDNALKYCTERKQFGRELATFQLVQERLMRVLGM LQGVLLLVLRLGRLMDEGKPHMMAHIALAKATCSRVAREGIALCREVLGGNGIVTDFGVA KFHADIEALYTYEGTYDINMLIAGRAVTGRSAFK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_231900 | 213 S | GLRTSAKKV | 0.995 | unsp | TGME49_231900 | 213 S | GLRTSAKKV | 0.995 | unsp | TGME49_231900 | 213 S | GLRTSAKKV | 0.995 | unsp | TGME49_231900 | 45 S | NDNSSDELQ | 0.992 | unsp | TGME49_231900 | 80 T | DSLLTPRER | 0.992 | unsp |