

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
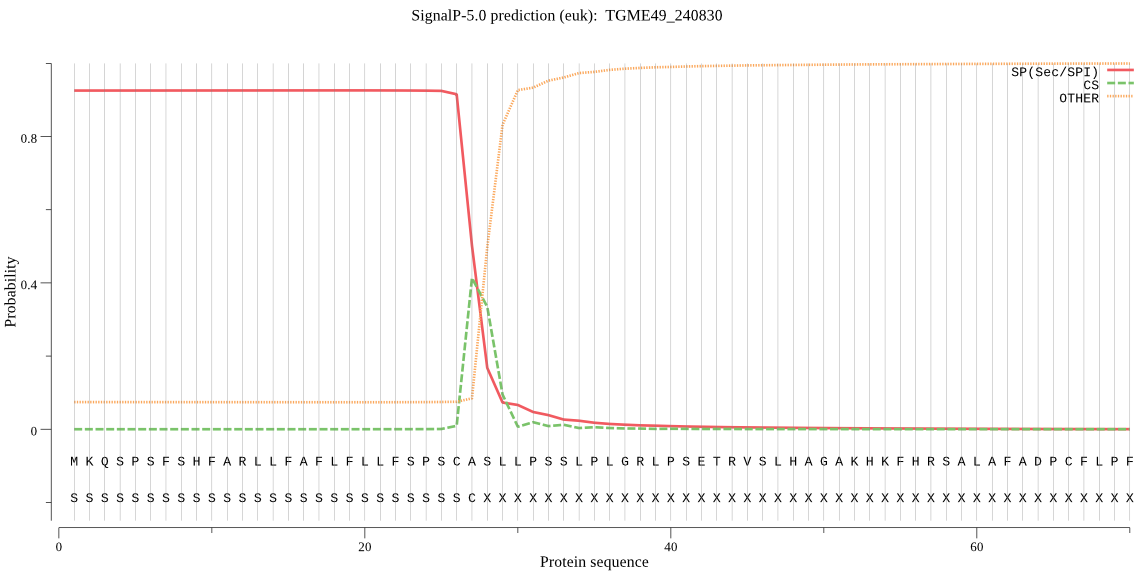
| TGME49_240830 | SP | 0.100845 | 0.899069 | 0.000087 | CS pos: 27-28. SCA-SL. Pr: 0.3915 |
MKQSPSFSHFARLLFAFLFLLFSPSCASLLPSSLPLGRLPSETRVSLHAGAKHKFHRSAL AFADPCFLPFLAPRSETVFRLPNVSTLLRDTPKMRLSACSSLPRSLSSAFVVPLSSLSSL SSLSSLFPLSSPSPLSSCFSRARLSREVDWSCSVRRAGQRDSSEPRNHATPLQSVPVSSL SHSLCSPSFARAGISSLRATEGCRRAEEAADQEREESSAFFSADAPALSFEVLQSRAKPV REDAPALLVVHGLLGSRRNMRSFSALLNSPKIVAVDLRNHGDSPWRDQMRVSDLGRDLLY MLHSKPDLFSSSALASSSSLLSAPRDVVLVGHSLGGLAAMYAALRAEEASRGRDALPRVK GLVVLDVAPVDYSGSRQAQQPVSSQTVVNLLCDLPMSAFEDRRQLERTLGATDPPLPRAM IQWLMTAVRERREKKPLDGGSIAWRAAGRPSRTADKTLKKDEKIRLEWRMNLLAIKQMLK TKQLRWPSEEFDAERRRKSRVSGCRDTAADAPVDAEGELGRSTAAEGEQRAAHAFEGPAL FLRGSNSQYVDVKRDWDTILRYFPNAEHRTVQNAGHWLHAEQPVQTAELINQFLAKV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_240830 | 499 S | RRRKSRVSG | 0.997 | unsp | TGME49_240830 | 499 S | RRRKSRVSG | 0.997 | unsp | TGME49_240830 | 499 S | RRRKSRVSG | 0.997 | unsp | TGME49_240830 | 502 S | KSRVSGCRD | 0.998 | unsp | TGME49_240830 | 549 Y | SNSQYVDVK | 0.991 | unsp | TGME49_240830 | 145 S | RARLSREVD | 0.991 | unsp | TGME49_240830 | 163 S | QRDSSEPRN | 0.994 | unsp |