

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_246550 | SP | 0.035475 | 0.964411 | 0.000113 | CS pos: 31-32. ANA-LI. Pr: 0.7996 |
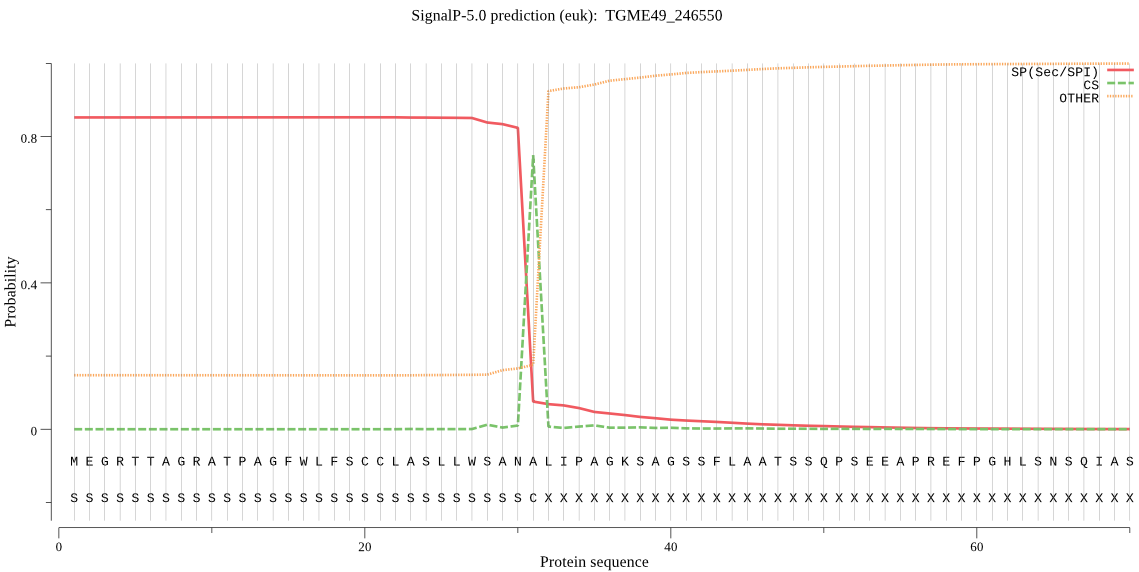
MEGRTTAGRATPAGFWLFSCCLASLLWSANALIPAGKSAGSSFLAATSSQPSEEAPREFP GHLSNSQIASETGSALKIEEIASSGSAPEVSGAAGASASKTSEKPIRPYHTGPSSRSSAY SASSGIPLCHTCSSFDAANGGCRLCVHGEVASDFLMPMMPIRNIDTHREETLSRLEANHR THLNEKKNFYVLRGKGPFGSLGNLPGTPGLDAAIAFGLSSPDLPSASFAQIKNKDSSDSG DVAAVGEETDSAVADGSKTLDLDLKLAETSVPILQMKDSQYVGVIGIGTPPQFVQPIFDT GSTNLWVVGSKCTDDTCTKVTRFDPSASKTFRAANPPVHLDITFGTGRIEGSTGIDDFTV GPFLVKGQSFGLVESEGGHNMHGNIFKTINFEGIVGLAFPEMSSTGTVPIYDNIISQGTL KENEFAFYMAKGSQVSALFFGGVDPRFYEAPIHMFPVTREHYWETSLDAIYIGDKKFCCE EGTKNYVILDSGTSFNTMPSGELGKLLDMIPSKECNLDDPEFTSDFPTITYVIGGVKFPL TPEQYLVRSKKNECKPAYMQIDVPSQFGHAYILGSVAFMRHYYTVFRRSDGTRPSLVGIA RAVHNDDNSAYLSNVLNEYPGAHIRKEDLMMERSMSAPSMREL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_246550 | 118 S | SSRSSAYSA | 0.996 | unsp | TGME49_246550 | 118 S | SSRSSAYSA | 0.996 | unsp | TGME49_246550 | 118 S | SSRSSAYSA | 0.996 | unsp | TGME49_246550 | 166 T | RNIDTHREE | 0.991 | unsp | TGME49_246550 | 237 S | NKDSSDSGD | 0.993 | unsp | TGME49_246550 | 639 S | MSAPSMREL | 0.996 | unsp | TGME49_246550 | 52 S | SSQPSEEAP | 0.997 | unsp | TGME49_246550 | 102 S | ASKTSEKPI | 0.995 | unsp |