

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
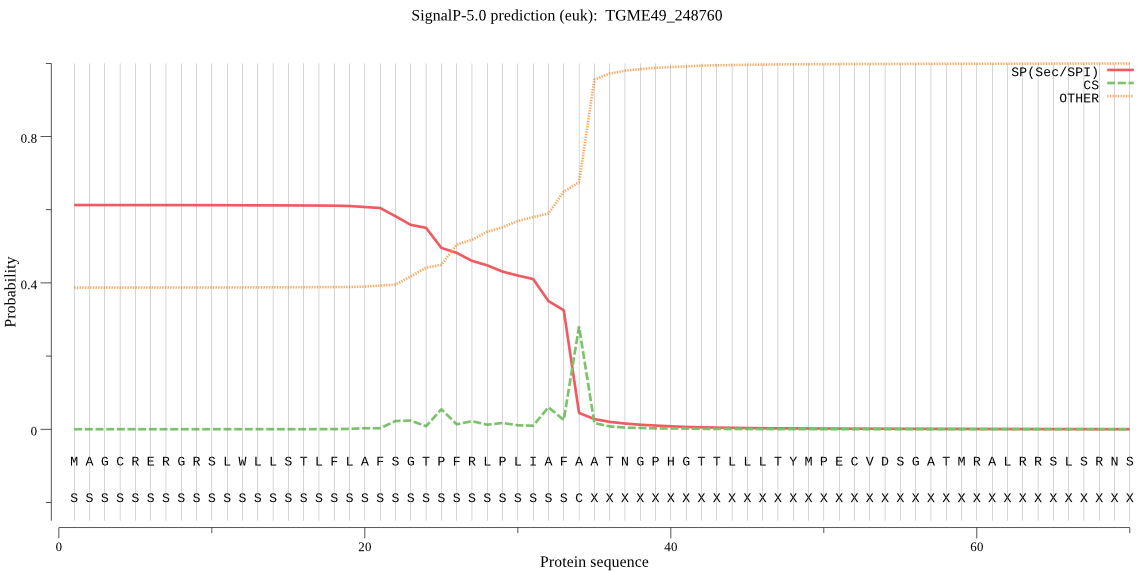
| TGME49_248760 | SP | 0.152512 | 0.839005 | 0.008483 | CS pos: 34-35. AFA-AT. Pr: 0.4193 |
MAGCRERGRSLWLLSTLFLAFSGTPFRLPLIAFAATNGPHGTTLLLTYMPECVDSGATMR ALRRSLSRNSAYKASHKSGLRMNESAHEDIEAIPLTARVIDEYAAYRTKVISLDSAKQPE QVGEDFEPVRCSISEESLKRVGVDVIELRNCEMVGGDEMKEALETDPCVASVEYDEELSI DSLQFPAAERDTSEDDPHANSPSPSSDRFSDRRRIATTSPSAANSIGEPLVSVDAPADGG AGYWRDKSGLNDSLYDLADCDPKRVVAVIDTGVSYTHPALVKNMWVNQNEISGNGIDDDS NGFIDDVYGFNFRENKGDPMDDHGHGTHVAGIIGAIKDQNSRVKGVCGNTSIAALKFMGA NGNGSTSDAIKALNYAVQMGIPLSCNSWGGPTWSEALIAALEAAESVGHLFIAAAGNQGR NTDEIPHYPASYRLANVVSVAATNSEDQLAPFSNRGAATVDLAAPGVKILSTFPPDQFRE LSGTSMATPVVAGVAAILMSLPYKDTKQIKTALVNGVDKMPATQGFVKSGGRVNASRSLS WLALELDLGEDMKARRKETEIEVNDTEVEEGRIEHSLAVEPVAA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_248760 | 193 S | ERDTSEDDP | 0.998 | unsp | TGME49_248760 | 193 S | ERDTSEDDP | 0.998 | unsp | TGME49_248760 | 193 S | ERDTSEDDP | 0.998 | unsp | TGME49_248760 | 206 S | PSPSSDRFS | 0.994 | unsp | TGME49_248760 | 210 S | SDRFSDRRR | 0.997 | unsp | TGME49_248760 | 365 S | NGNGSTSDA | 0.993 | unsp | TGME49_248760 | 65 S | ALRRSLSRN | 0.995 | unsp | TGME49_248760 | 70 S | LSRNSAYKA | 0.99 | unsp |