

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
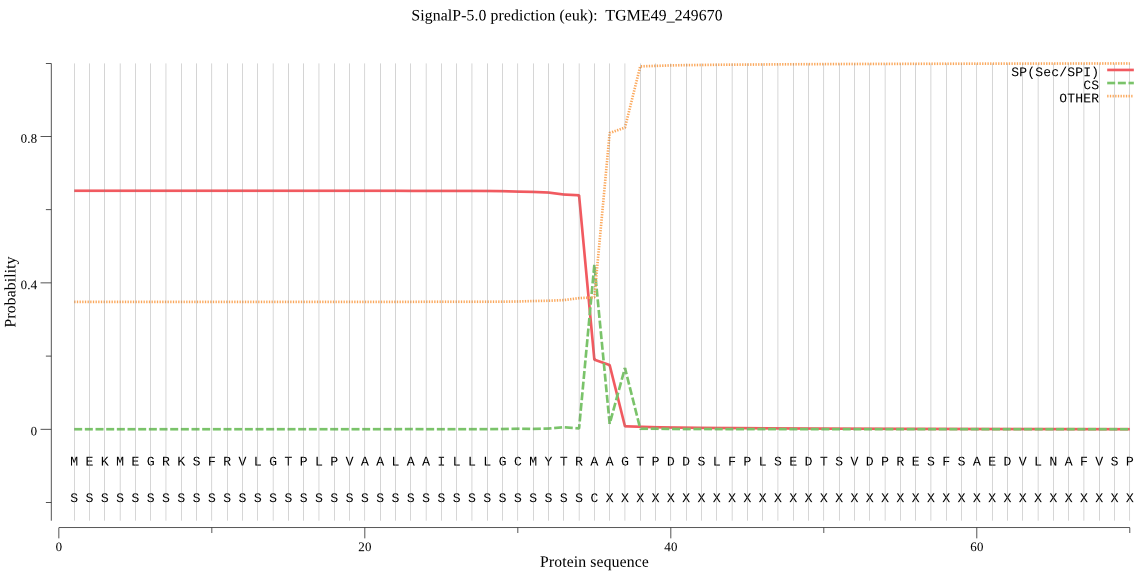
| TGME49_249670 | SP | 0.138308 | 0.861344 | 0.000348 | CS pos: 35-36. TRA-AG. Pr: 0.4969 |
MEKMEGRKSFRVLGTPLPVAALAAILLLGCMYTRAAGTPDDSLFPLSEDTSVDPRESFSA EDVLNAFVSPESVESLFASIVAEQVVATSGNLTESAPRDRDSAPERRSDGGTLRGHPEDA GELLRLLLADSEDMELWKANFFRHLTHSMRHIVRDSVLVSEKAFPSEQADEAGRDPGDLK KALKETGEDVFWESRPASSNAAVALIKRKMEKQAGKTGEGAGHTWEPEVSLRFRYLSLKD AKKLMGTFLVNTKVEGFPTPKGMPLPAKEFENATEPVPAHFDARTAFPACKDVVGHVRDQ GDCGSCWAFASTEAFNDRLCIRSQGKGLMPLSAQHTTSCCNAIHCASFGCNGGQPGMAWR WFERKGVVTGGDFDALGKGTTCWPYEVPFCAHHAKAPFPDCDATLVPRKTPKCRKDCEEQ AYADNVHPFDQDTHKATSAYSLRSRDDVKRDMMTHGPVSGAFMVYEDFLSYKSGVYKHVS GLPVGGHAIKIIGWGTENGEEYWHAVNSWNTYWGDGGQFKIAMGQCGIDGEMVAGEAAWQ ETEGVVNGEEELPGQHASGARVGAHAEEEREM
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_249670 | 72 S | VSPESVESL | 0.995 | unsp | TGME49_249670 | 72 S | VSPESVESL | 0.995 | unsp | TGME49_249670 | 72 S | VSPESVESL | 0.995 | unsp | TGME49_249670 | 102 S | RDRDSAPER | 0.996 | unsp | TGME49_249670 | 108 S | PERRSDGGT | 0.993 | unsp | TGME49_249670 | 237 S | FRYLSLKDA | 0.998 | unsp | TGME49_249670 | 410 T | VPRKTPKCR | 0.992 | unsp | TGME49_249670 | 438 S | HKATSAYSL | 0.992 | unsp | TGME49_249670 | 9 S | EGRKSFRVL | 0.991 | unsp | TGME49_249670 | 59 S | RESFSAEDV | 0.995 | unsp |