

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
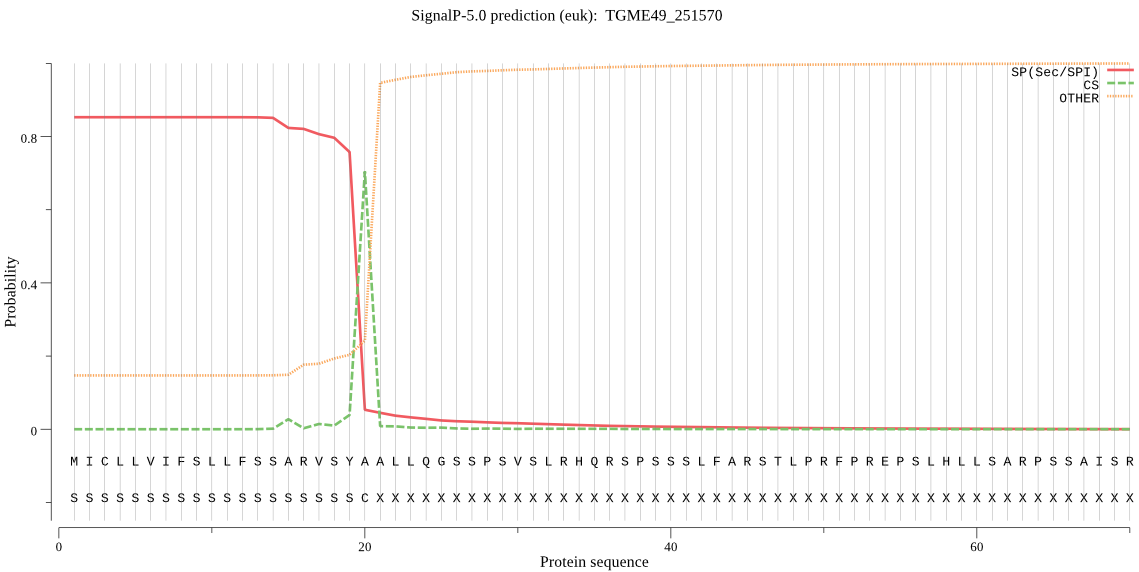
| TGME49_251570 | SP | 0.017243 | 0.968976 | 0.013781 | CS pos: 20-21. SYA-AL. Pr: 0.8879 |
MICLLVIFSLLFSSARVSYAALLQGSSPSVSLRHQRSPSSSLFARSTLPRFPREPSLHLL SARPSSAISRFSSSSVASLSPRPPPPQSQARCPASFPSASSPSSCFPPPPPSSSPRLAGR FGLQHAPPVAFGAFPASRKAAISADAGCRGAATLERRSEEAAHAFRSPGRLPRSAPACFS ASLKLQVSGAASPPRCLDTPVFSATSNHPWEDGATRRRAASSLGFPRSSASSPSSASSLS EFAVLRDPVPIADFENGADKREKLGRPGESPDLREKRSRALAFQSLPPASSHPAASLPLR KDADVSLSSSASLHSGHALPERQADSSASSEPSRSSEPSPSCVEGIDSSTSPPGPAAASS RPPASAQDRGKTPCQSSSPSRAPSARGLPCRLAAFLPPSVSLLFSRLRPRPPHSSPFSFP LTSSLSSSPLCSPLFASSSAYVSRAPRPPSPLLCSRASSSPPSGREGPLWQRWLHAFSSG RPWPSSKTHGWSSSQTPASESLSDASCSSLPSSSPSSSSPSSSSVSATAEREAGQFLQRL ERFRESARRLFRLGVSRCFAFGSALNSRLRESVLAQAAVVLAVYFVHFAYISQQTFVLPV QLLPNRCGLFQHVQGDSLAGWAALALLCASKSQAFSNLANEKSKSTRAAIQRPYTPSALS SSSSPFSSPSSQPSSWSSPSSSPPSLFSASLSSVSPSGVSTPRGQVTEAASSRSGGAEVP PCAHASGTTDLFVGASAAEEDDTERQTKERRRLLPGGRHAQVSRKLPWHGPFPAFLKSAS LLLALLGSYVLSGYVACGLDFLFYFLHAAGVVTLDVAMHRALQVLLAHLAWVFMGVALLR LSPITFFQSAWEPPHYEAPAPACSETAREETEEEPTGSPPFQSDSTEGKRDGEEPQDQTR RLGAGEGKDEETVDACEERSAGLQVEQRSCATLSSRWRKGPVASLLLSRDTDRESNAGLP ADGNESSNSKRVHDAKLPPRQLEGCFFLASRRGEEAGLGGMVEAETEKHLLSSGNKVAGS CEFFRRLWWRIKRPREAERNFDTQARHRESLRSPHSGLKPQDALQTSARVGCTYTPGLGA SGSHAKAKTGSSLVEKRANARWFTVRSRGKGGLWVWPVISGYLISCLFFNLTEFLNDALM SFLPEEPQGPTIVQHIMNPTLNTHLSFLVGALAPCLSAPGWEELLYRGFCLPLFSQVMPL SLAAVLSSLLFAVHHMNVQTVLPLWVLGLTWTAVYVHSKNLLTTVLIHAMWNSRIFLGNL FGC
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_251570 | 232 S | SSASSPSSA | 0.997 | unsp | TGME49_251570 | 232 S | SSASSPSSA | 0.997 | unsp | TGME49_251570 | 232 S | SSASSPSSA | 0.997 | unsp | TGME49_251570 | 238 S | SSASSLSEF | 0.993 | unsp | TGME49_251570 | 329 S | DSSASSEPS | 0.994 | unsp | TGME49_251570 | 335 S | EPSRSSEPS | 0.993 | unsp | TGME49_251570 | 339 S | SSEPSPSCV | 0.993 | unsp | TGME49_251570 | 351 S | DSSTSPPGP | 0.992 | unsp | TGME49_251570 | 384 S | SRAPSARGL | 0.995 | unsp | TGME49_251570 | 463 S | SSPPSGREG | 0.998 | unsp | TGME49_251570 | 501 S | PASESLSDA | 0.996 | unsp | TGME49_251570 | 519 S | PSSSSPSSS | 0.995 | unsp | TGME49_251570 | 546 S | RFRESARRL | 0.996 | unsp | TGME49_251570 | 951 T | LSRDTDRES | 0.99 | unsp | TGME49_251570 | 1050 S | RHRESLRSP | 0.995 | unsp | TGME49_251570 | 1053 S | ESLRSPHSG | 0.993 | unsp | TGME49_251570 | 80 S | VASLSPRPP | 0.994 | unsp | TGME49_251570 | 158 S | LERRSEEAA | 0.998 | unsp |