

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
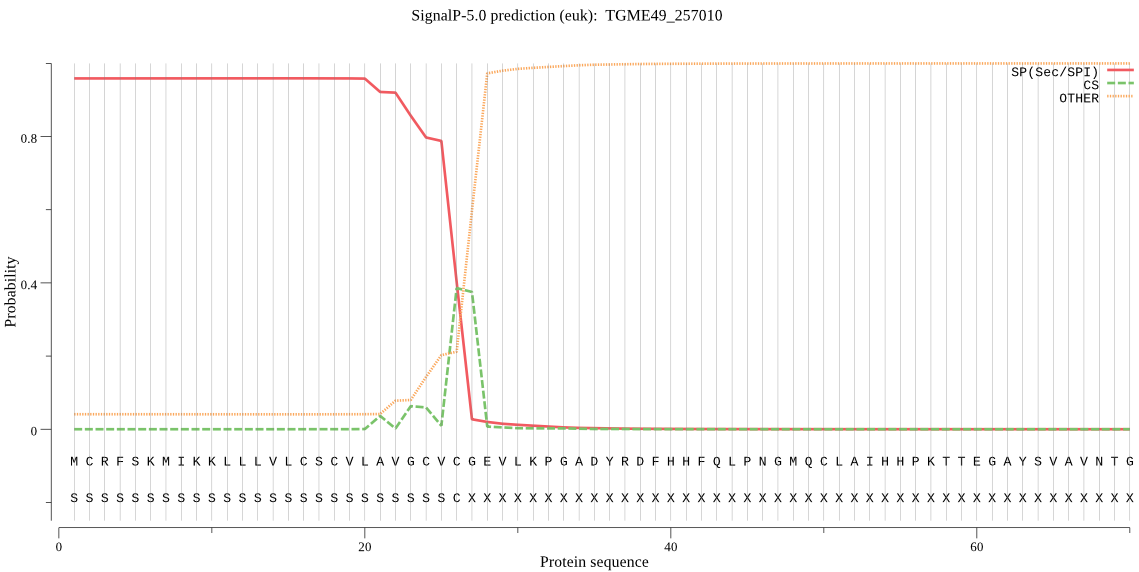
| TGME49_257010 | SP | 0.006471 | 0.993490 | 0.000039 | CS pos: 27-28. VCG-EV. Pr: 0.4959 |
MCRFSKMIKKLLLVLCSCVLAVGCVCGEVLKPGADYRDFHHFQLPNGMQCLAIHHPKTTE GAYSVAVNTGSLYDPEDLPGLAHFLEHMLFLGTSKHPEPESYDKFMSERGGQNNAYTDEE KTVFFNQVSDKYLEDALDRFSQFFKSPLFNPEYEEREAHAVDSEHQKNVPNDEERTWFTI RSLAKGPLSRFATGNLETLNTAPKRKGINVVSRLKDFHKKYYCASNMAVVIMSPRSLVEQ ETLLRKSFEDVTSGNPNFLGFDQCPGVDYDRTPPFDLSNTGKFIHLQSVGGESSLWVAFS LPPTITSYKKQPTGILTYLFEYSGDGSLSKRLRTMGLADEVSVVADRTSVSTLFAVKVDL ASKGASERGAVLEEVFSYINLLKNEGVDSKTISSISEQSLVDFHTSQPDPPAMNEVARLA HNLLTYEPYHVLAGDSLLVDPDAQFVNQLLDKMTSDHAIIAFADPQFKRNNDSFDVEPFY GIEYKITNLPKEQRRRLETVTPSPGAYKIPPALKHVPRPEDLHLLPALGGMSIPELLGDS NTSGGHAVWWQGQGTLPVPRVHANIKARTQRSRTNMASRTQATLLMAALAEQLDEETVDL KQCGISHSVGVSGDGLLLAFAAYTPKQLRQVMAVVASKIQDPQVEQDRFDRIKQRMIEEL EDSASQVAYEHAIAAASVLLRNDANSRKDLLRLLKSSSTSLNETLKTFRDLKAVHADAFI MGNIDKADANSVVQSFLQDSGFTQIPMKDAAQSLVVDQRAPIEALIANPIPKDVNHATVV QYQLGVPSIEERVNLAVLGQMLNRRLFDRLRTEEQLGYIVGARSYIDSSVESLRCVLEGS RKHPDEIADLIDKELWKMNDHLQSISDGELDHWKESARAELEKPTETFYEEFGRSWGQIA NHGHCFNKRDLELIYLNTEFNRKQLSRTYTKLLEPTRRLVVKLVASLQPAQEVTLIGQES LLDERRESLAHKRSKGQKHRKGLTLENLDEETVSQFHNAAGFYPQVTVCELSPRMLLRLE EEM
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_257010 | 366 S | SKGASERGA | 0.991 | unsp | TGME49_257010 | 366 S | SKGASERGA | 0.991 | unsp | TGME49_257010 | 366 S | SKGASERGA | 0.991 | unsp | TGME49_257010 | 686 S | NDANSRKDL | 0.998 | unsp | TGME49_257010 | 700 S | SSSTSLNET | 0.995 | unsp | TGME49_257010 | 824 S | VGARSYIDS | 0.997 | unsp | TGME49_257010 | 829 S | YIDSSVESL | 0.991 | unsp | TGME49_257010 | 968 S | ERRESLAHK | 0.994 | unsp | TGME49_257010 | 236 S | MSPRSLVEQ | 0.996 | unsp | TGME49_257010 | 247 S | LLRKSFEDV | 0.998 | unsp |