

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
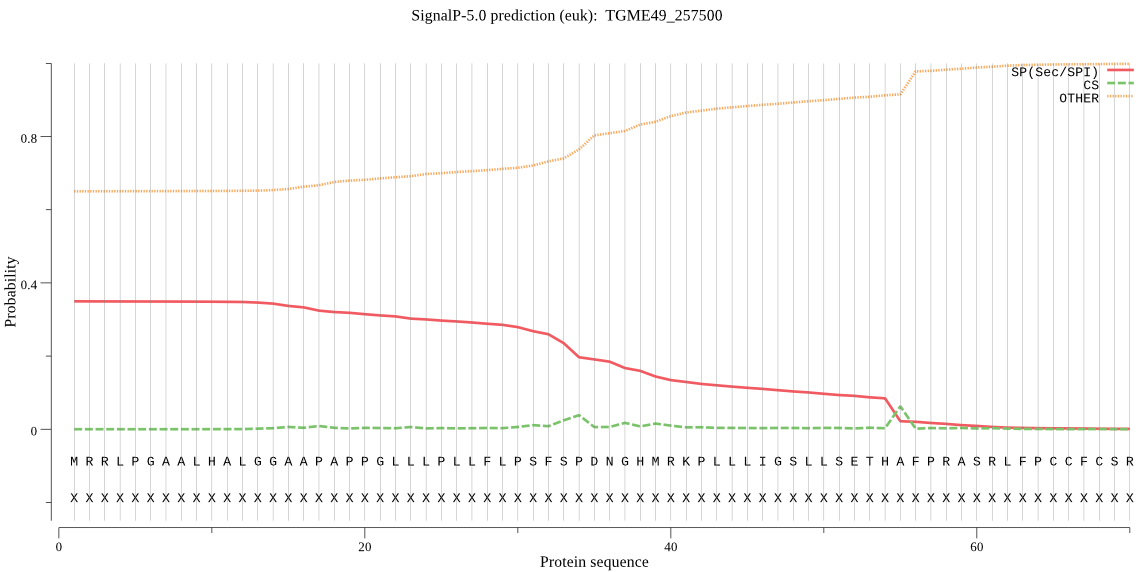
| TGME49_257500 | OTHER | 0.631341 | 0.366442 | 0.002218 |
MRRLPGAALHALGGAAPAPPGLLLPLLFLPSFSPDNGHMRKPLLLIGSLLSETHAFPRAS RLFPCCFCSRFSASSELGSEAFGVACLACGPPAASQLTSSCAACLATSLRALSPCGSSPS LSRPAVSFRPEGTRCYSSVSSRGHWQPRSETQVSTASFASLRRIARGGQRIVARRRPASE LCRGGLRGRSTTVTGLSPGERVGGAREKTKETCAKSRDSLDQASPRFWAIRQPQQPLASR LSNMLAASGCLFSSASPLSFSSPSSLSSPPFICSPSRVCSSERDPSFQRAGLETSACSPH RKRCGRSGDMARRQERRFCRFRPGDTRGLGRTSLAACDSGGFKAVSPQAGAETDESDSCA KESREGKAIHLQRASRAEAEEPSEAGGKPRGATVSTSLQRLYRHDGWPEEGELVNGQGLA LRTYTWWPCQCNPCLASLGRASSKYTDASLRLEDPASLPSSFLSSSCSSVSPPSSSSASS LLGKLPTARWWLRGSESAAAAGPVPSREEEGQANTEETVQRSSAEKKREGNGDWWCLVGE KGELQRDRKTGRTACAACNKPIRGVVILVHGYGEHCRSHFLQKLQERPRASSAPTETTEE EGRRGVGGDPVEAERDADFNRASDKRSSLPDSPPATASVSSDSSVSSSSFSSRASSLDLS DASSDLPSVFAPGPESSLGSLFYEGSWVHALNARGFLVVGFDLQGHGLSGAWRGLRCVVS ELDDFARDALLVVLSTQRRFGRPDRDAFPFHLLGISMGGWTAARAVELAGHSETLSRCWA HVGATPETPHKRARETGEEDSEGRGATPAAMRGEEKAGGEELDRCGRSALLGAAGREGER KTRKERESGEDKDEQQLASKKTPRANLSPGSHQIPSEVANLALGLTGLILVSPMFDLERR KAKLKWELAKYGILPLAAFFPGVPLELFAPWRRLKSDRKRKRVAEYEKLRLSFQADPLTF KASPPGGLVAAIMRGAQRALEPEEVEKINCENVERVLILHNATDSICDAGGAVQFFQRLG SGENEDAQTSGEEETRECGEAERFRQGDPDAARRPAKRRGQKAPEKALILLNVGAAERSS GKREDNLTEQQRSFAAFVERERARAALENSQSGEDAALSKAAGGGQAGGNRGANALCESA EDAEVRGTRGKGQKVRRSGAEAERDSTEQVEKARKKEKVSFVENVDVWHNLANEPGQEKV FELLDAWLEGDTQRKECAARKEQTEPAERGRRPEE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_257500 | 219 S | KSRDSLDQA | 0.994 | unsp | TGME49_257500 | 219 S | KSRDSLDQA | 0.994 | unsp | TGME49_257500 | 219 S | KSRDSLDQA | 0.994 | unsp | TGME49_257500 | 280 S | SRVCSSERD | 0.994 | unsp | TGME49_257500 | 281 S | RVCSSERDP | 0.996 | unsp | TGME49_257500 | 442 S | LGRASSKYT | 0.997 | unsp | TGME49_257500 | 523 S | VQRSSAEKK | 0.997 | unsp | TGME49_257500 | 623 S | FNRASDKRS | 0.996 | unsp | TGME49_257500 | 628 S | DKRSSLPDS | 0.998 | unsp | TGME49_257500 | 632 S | SLPDSPPAT | 0.992 | unsp | TGME49_257500 | 651 S | SSSFSSRAS | 0.992 | unsp | TGME49_257500 | 655 S | SSRASSLDL | 0.997 | unsp | TGME49_257500 | 656 S | SRASSLDLS | 0.991 | unsp | TGME49_257500 | 848 S | KERESGEDK | 0.998 | unsp | TGME49_257500 | 1021 S | QRLGSGENE | 0.996 | unsp | TGME49_257500 | 1030 S | DAQTSGEEE | 0.996 | unsp | TGME49_257500 | 1080 S | AERSSGKRE | 0.998 | unsp | TGME49_257500 | 1112 S | ENSQSGEDA | 0.993 | unsp | TGME49_257500 | 1158 S | KVRRSGAEA | 0.997 | unsp | TGME49_257500 | 1166 S | AERDSTEQV | 0.996 | unsp | TGME49_257500 | 1180 S | KEKVSFVEN | 0.992 | unsp | TGME49_257500 | 113 S | LRALSPCGS | 0.996 | unsp | TGME49_257500 | 197 S | VTGLSPGER | 0.996 | unsp |