

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_262920 | SP | 0.018191 | 0.981324 | 0.000485 | CS pos: 31-32. VSS-WI. Pr: 0.5532 |
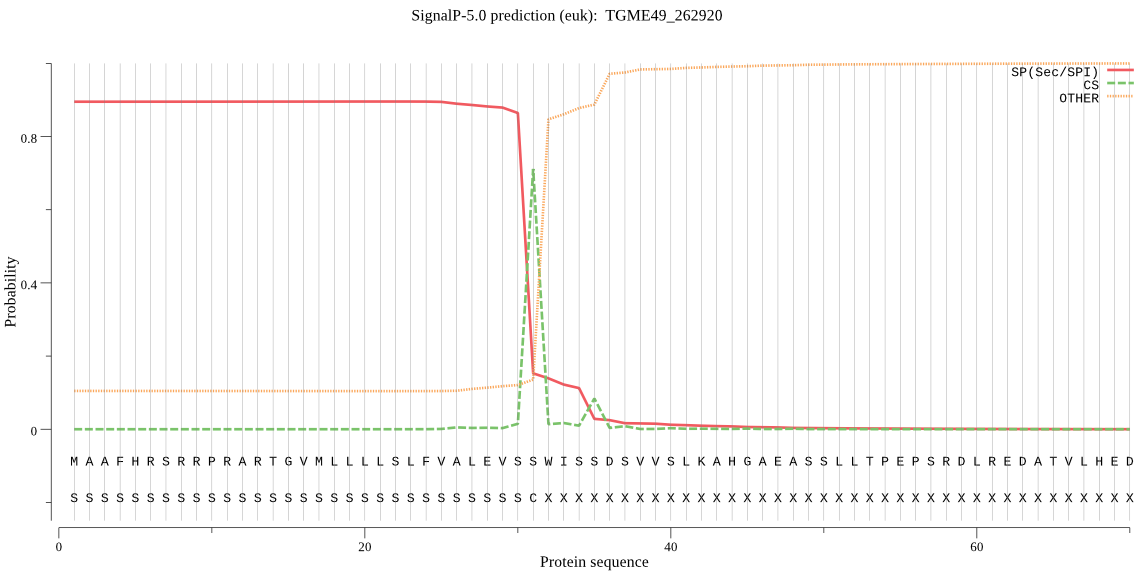
MAAFHRSRRPRARTGVMLLLLSLFVALEVSSWISSDSVVSLKAHGAEASSLLTPEPSRDL REDATVLHEDGFVSTFPAAVATSAHGAEPLPPEREITEEVTGEMLVGSSLLQVSAAGQPT ELAAAAAEGVNSQGGNTLAKTLAAAAASQKQTTAKDRHALLTSSLSSVVKIFVDITMPDY FSPWQMQAPKDASGSGFVVEGKRILTNGHVVGETTRVLVRKHGNAKKFLARVVATAHEAD LALLEVESDEFWENLQPLPFGGIPRLRDSVTVLGYPTGGDQLSITEGIVSRVGMSMYAHS SVSLLTVQIDAAINPGNSGGPALVDGRVVGVAFQGFSHLQNVGYIVPYPIIEHFLNDLVL HGRYTGFPSLGVKVSHMENDHLRQFKGLSALKASDLPPGVTPTGVLVVEVDNLRVSRYKA GKIRVPYTSRTLSGPRNLKMMQSVQAQEDVSLPSATSSGHGASFSGSLASPVPDFVKEAA PPGQMLSAPLGGETPESAVVAAGGGEKPAARAFGQGGSEAGPSRSQPTFLQTQVTPKHIL ANRSQLLRLYALARRRQLTREAVADEAAADSGEERGLVETQGARTTAPPVMVRMVVRKLG PGIYSQIPEGDRSFRAREGSGFPKPWSAFRMLQRRLRNVQVTARGEKTGGEDETGDLSQD QEAKGNREEPEQVEGKVLVSPENVPGSIQKTGALPVNMRGPLLSTALPGSSSISSFPAQE PLSEKASEAVEEGPRSEEELLLPNPYFHQQDAEENEIGFKVGDVILAIDGIDVADDGTVA FRQLERVSIDYTIMKRFNGETCKALVLRDGQVREVLLPITNLNLKVPAHTWDQKPKYFVF GGLVFTTLTRHLLEHMKLTEFPAEFFTKIKQTKYQEEEGDEVVVLSVILASELTVGYTAA PAIVTAVQGQKVRGLADVVRIVEQSTDNFLEFTVKISGISALPIVLDRKKAMAVNPKILG QHKILRDRSYFL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_262920 | 571 S | AAADSGEER | 0.991 | unsp | TGME49_262920 | 571 S | AAADSGEER | 0.991 | unsp | TGME49_262920 | 571 S | AAADSGEER | 0.991 | unsp | TGME49_262920 | 680 S | KVLVSPENV | 0.992 | unsp | TGME49_262920 | 723 S | QEPLSEKAS | 0.997 | unsp | TGME49_262920 | 736 S | EGPRSEEEL | 0.997 | unsp | TGME49_262920 | 416 S | NLRVSRYKA | 0.992 | unsp | TGME49_262920 | 433 S | SRTLSGPRN | 0.993 | unsp |