

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_263290 | OTHER | 0.999901 | 0.000006 | 0.000093 |
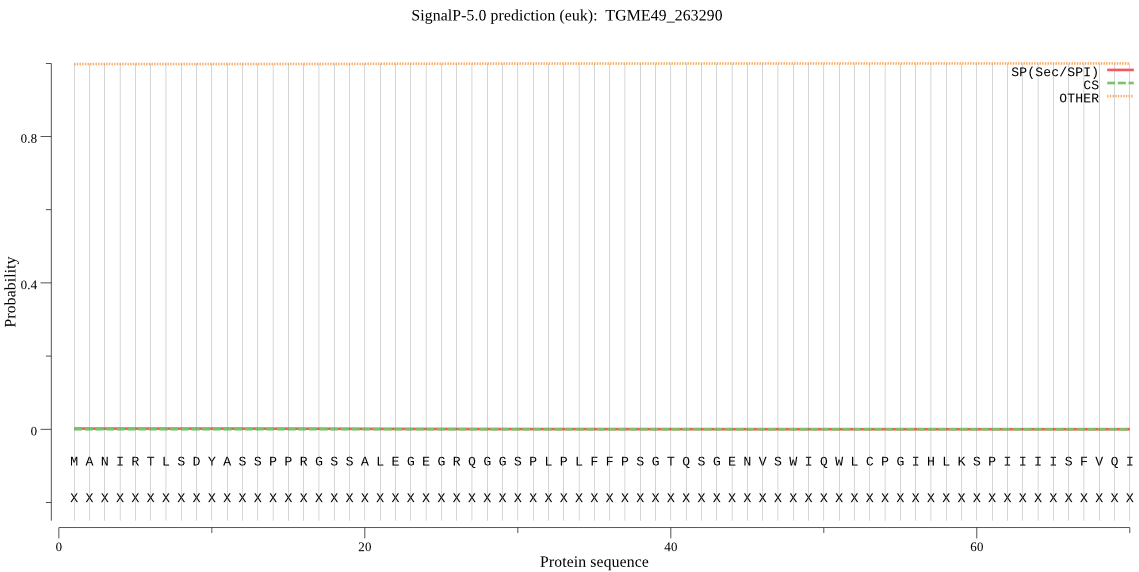
MANIRTLSDYASSPPRGSSALEGEGRQGGSPLPLFFPSGTQSGENVSWIQWLCPGIHLKS PIIIISFVQIAVYIASLAAGLAPNEILAPTPQTLVMFGANIPELIRVGEIWRLICPLFLH LNLFHILMNLWVQIRIGLTIEEKYGWKMLLAVYFGVGVLANMISAAVLFCGQMKAGASTA VFALIGVQLAELALIWHAIQDRNSAIISVCICLFFVFVSSFGSQMDSVGHIGGLVMGFAA GIWLNENSDIKPTWYDRARLTSQVALAAAPILSCIFIFLVPRC
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| TGME49_263290 | 19 S | PRGSSALEG | 0.992 | unsp |