

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_263470 | OTHER | 0.999972 | 0.000021 | 0.000007 |
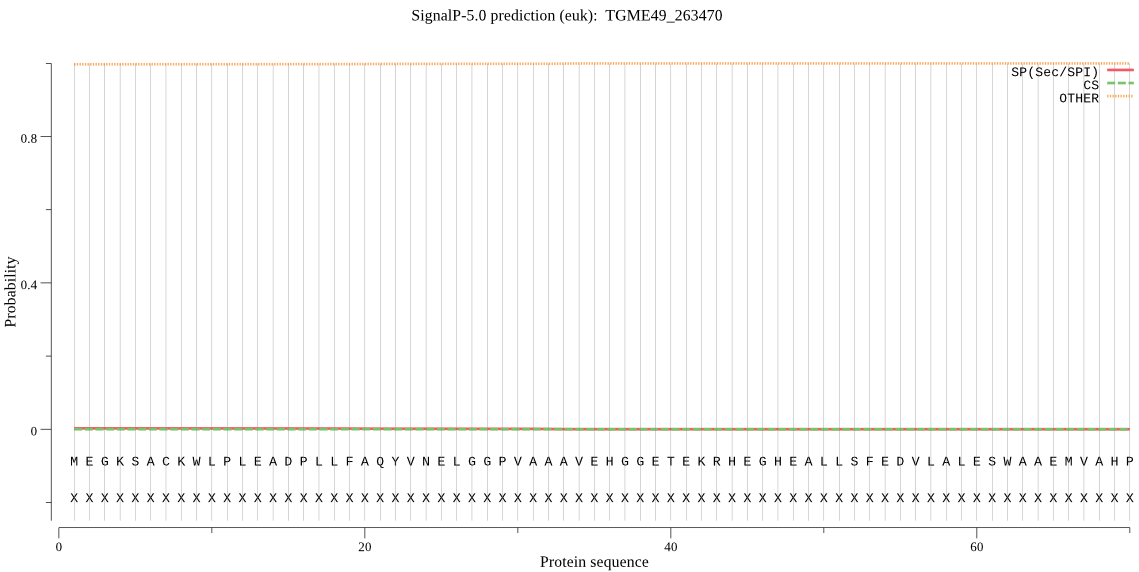
MEGKSACKWLPLEADPLLFAQYVNELGGPVAAAVEHGGETEKRHEGHEALLSFEDVLALE SWAAEMVAHPTVAVLLLFPITEATEKGRREQDKQTAGQSLNNVWFTKQTVGNACGTVALL HCLANLPREKFPLQPNRFLEHFLKETADLSPEQRAKVLETDRSLASAHKSFEQQGQSAVP PRESDVDTHFVAFVFHEGHLVELDGRRATPVDHGSVEGGATLEDAARNQRLLKMTLNVIQ KEFVEKCPGELRFQVIAVGDAKAA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_263470 | 184 S | PPRESDVDT | 0.997 | unsp | TGME49_263470 | 209 T | GRRATPVDH | 0.992 | unsp |