

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
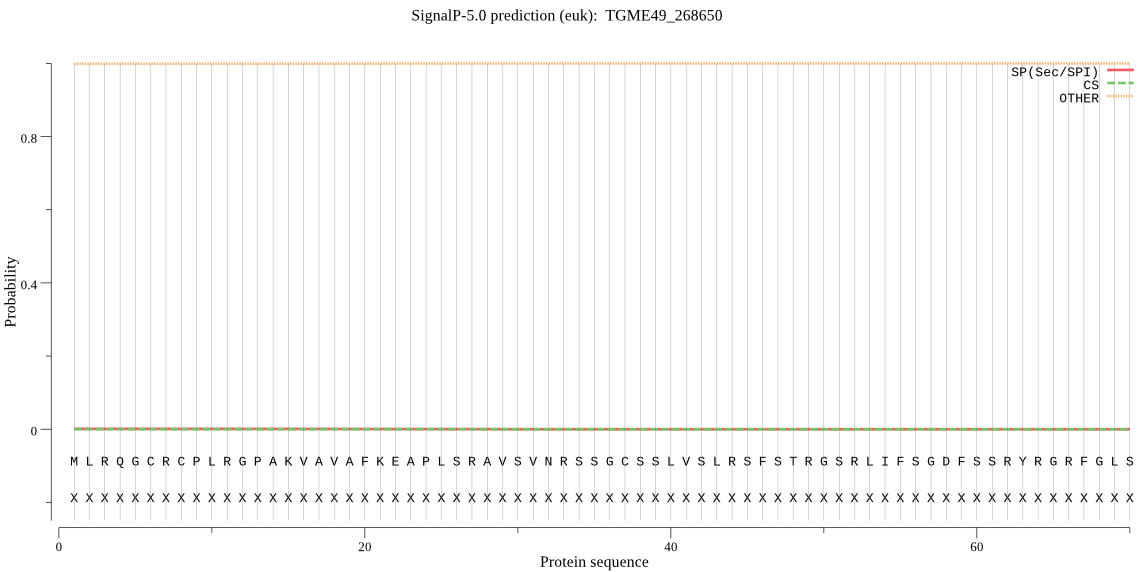
| TGME49_268650 | mTP | 0.221192 | 0.000111 | 0.778697 | CS pos: 46-47. RSF-ST. Pr: 0.2092 |
MLRQGCRCPLRGPAKVAVAFKEAPLSRAVSVNRSSGCSSLVSLRSFSTRGSRLIFSGDFS SRYRGRFGLSYSDLLGFERRPREALDHIPRFMQASCFSAAANTPGFSLKGDDYTDRALEA LAGMISLAQEYSPPTLETELLVLSLLNQGEDGLFTMIMKQAGVDLDKLRSGVVDFVERHP TVSDNSTRTLGPVLQKVLSSANSLRLQWHDEYISVEHLAAALADEDSRFLVRFLKESKLT ANDIRQAIKAIRGTRRVNTKTPEVSYQSLKKYGRDLTEAAMANELDPVIGRDKEVRRVIQ ILSRRTKNNPIILGDPGVGKTAIAEGLAQRIVSGDVPDTLAGRQLISLDLGALLAGAKLR GEFEERLKSVIREVQESSGQIILFIDEIHMVVGAGSAGESGMDAGNILKPMLARGELRCI GATTLDEYRKYIEKDKALERRFQVVLVDEPRVEDALSILRGLKERYEMHHGVSIRDSALV AACVLSNRYIQDRFLPDKAIDLIDEAASKIKIEVTSKPTRLDEIDRKLMQLEMEKISIVS DMKGGQDAAEQQRLQTLEKKMADLKEEQSRLNAIWEQERAEIEKIADLKQEIDEAKVEQQ KAEREYNLNKAAQIRYGKIPELTQKLATLEAAAKREELSSSRLLRDTVTAEDIAQVVGSW TGIPVSRLVEGEREKLLGLEKALNDRVIGQEEGVRSVAEAIQRSRAGLCDPNRPIASLVF LGPTGVGKTELCKALARQLFDTEEALLRFDMSEYMEKHSTARLIGAPPGYIGFDKGGQLT EAVRRRPYSVLLFDEMEKAHPEVFNIFLQILEDGILTDSHGHTVSFKNCIIIFTSNMGSD LLLQASGTKDREEVTQALMEIVRRHLRPELVNRMDEFVVFNPLSEANLFGIFDLEVAKLQ ARLTDRRLTLSVTHRAKADIVQAAYDPNFGARPLRRAVQHTLETPLSRLILSGSISNGRR VGVASKQEELPVSSEKIEDSKIPENLQFFLYPQEGTAQHMLASGDTGDATKDTGETAAFS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_268650 | 400 S | SAGESGMDA | 0.99 | unsp | TGME49_268650 | 400 S | SAGESGMDA | 0.99 | unsp | TGME49_268650 | 400 S | SAGESGMDA | 0.99 | unsp | TGME49_268650 | 473 S | HHGVSIRDS | 0.996 | unsp | TGME49_268650 | 909 T | DRRLTLSVT | 0.992 | unsp | TGME49_268650 | 47 S | LRSFSTRGS | 0.997 | unsp | TGME49_268650 | 369 S | ERLKSVIRE | 0.994 | unsp |