

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_269250 | OTHER | 0.999985 | 0.000011 | 0.000004 |
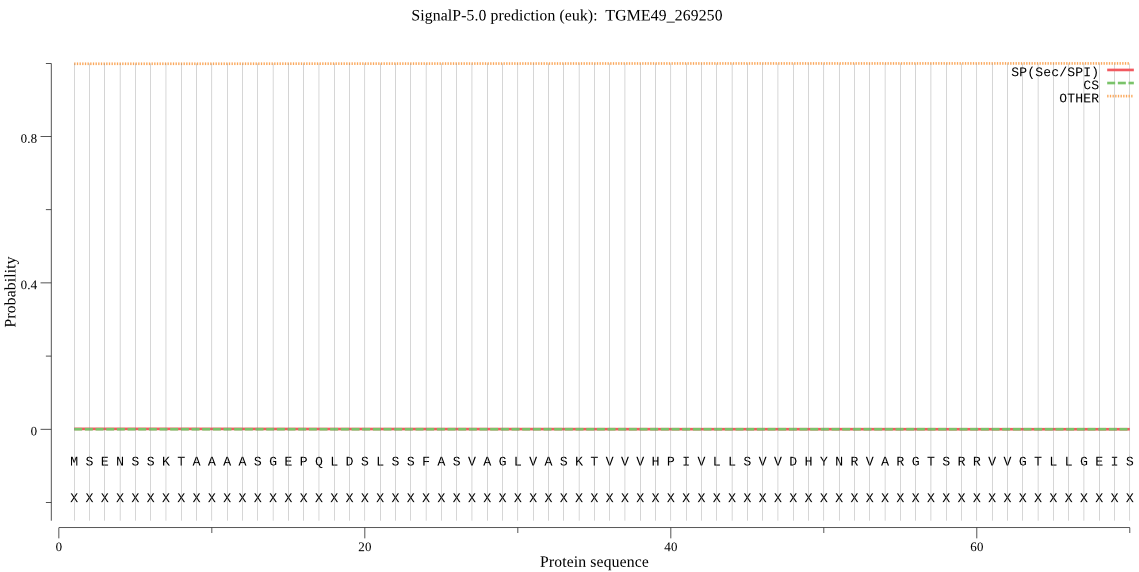
MSENSSKTAAAASGEPQLDSLSSFASVAGLVASKTVVVHPIVLLSVVDHYNRVARGTSRR VVGTLLGEISDGEIHVTNSFALPFEEDPKDPNVWYLDRNYHEHLYHMFKKVNTRERVLGW YSTGPQVRMTDLEIHEIFRRYTPNPVYVIVDINPKDSVVPTKAYYSFEQPTSDRTFRRTF VHVASTIGALEAEEVGVEHLLRDLKNASTSTLATRVADKLSALKLLIGKIQEIYAYLQDA ANKKIVANPNIMYTIQDIFNLLPDLSDPELIEAFTIQANDTMLNLYLGSVVRSVLALHNL INNKVENKRASEEKKEKKEDEETGETKKEAGTDTKKGEKESSQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_269250 | 311 S | NKRASEEKK | 0.998 | unsp | TGME49_269250 | 311 S | NKRASEEKK | 0.998 | unsp | TGME49_269250 | 311 S | NKRASEEKK | 0.998 | unsp | TGME49_269250 | 57 T | VARGTSRRV | 0.994 | unsp | TGME49_269250 | 157 S | NPKDSVVPT | 0.99 | unsp |