

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
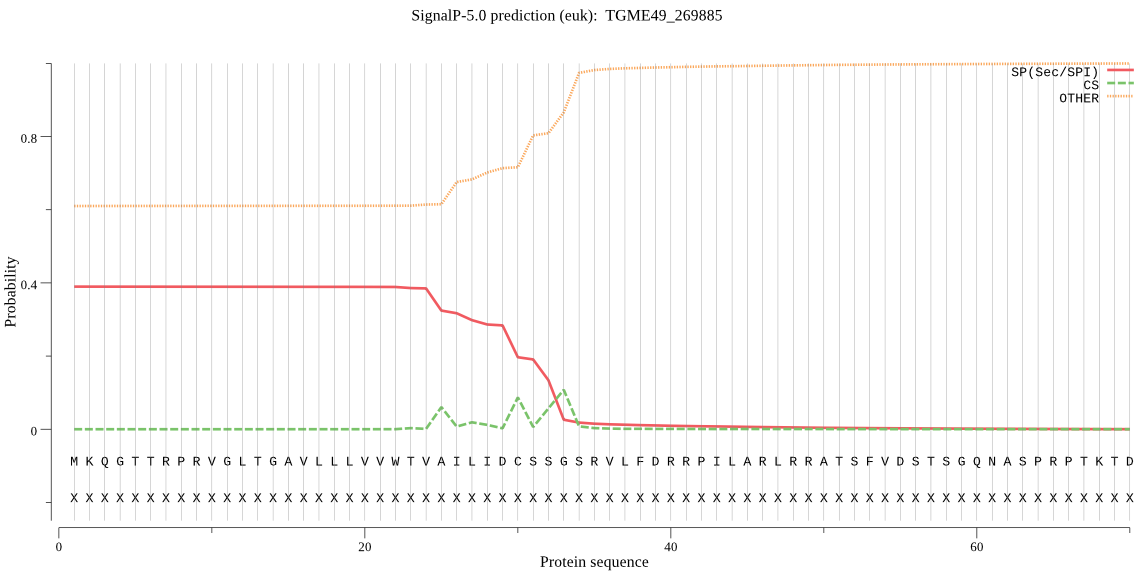
| TGME49_269885 | SP | 0.215228 | 0.782418 | 0.002353 | CS pos: 33-34. SSG-SR. Pr: 0.7198 |
MKQGTTRPRVGLTGAVLLLVVWTVAILIDCSSGSRVLFDRRPILARLRRATSFVDSTSGQ NASPRPTKTDKIRKPRNDSRNYRYIELPNELRALLVSDPECDEAAASMRVGVGSMSDPPK IPGLAHFTEHMLFQGSKRFPGTHDFFDFVHNHGGYTNAFTSKFSTVFSFSIGPGFLEPGL DRLADLFSAPLLKSENLLKEVNAVHSEYIIDLTDDGRRKHHLIRQTAKGGPFSNFTVGNL ESLMERTKQQGIDPVKAMREFHNKWYSSNLMTLAVVGRESLDVLESHVRKHFGNVPNGRV TPPVFEECSEAFIPLDPNELGTETLVVPEADLHDATFVFYLPPQAKNWRSKPLQFISEML EHEGPTSLSSKLKREGLITSLVTDYWSPELCTVLQVNVRLTEGGRSKESVYKIGHALFTF LRNLGVSRPERWRVTEMAKIRQLGFAFADMPDPYALTVRAVEGLNYYTPEEVIAGDRLIY HFDPDIIQQYVQKFLVPDNVRLFIFDKKLAADVDREEYWFKIKHGVEPIMDSAFKKWKEI TSAPANTVQSMMRMEGMALPAPNRFLPNNVAILPRPPGSGQGAEKSAFPEPLVFAGDHIC ANKCAVFHKQDTTFHSPKAVVELQIYYTGSHEDGVRERILTALYVQSLRFALRERFADTH RAGMSLGLGSGVAVGSTTLPTVKRQRVLTLSAAGFSDKLDYVLRAFAKSLAATEGASDEK PIPSPMELGVTMSHGVRHLMHSRVTAPSNVLASVQSSRVYTTAVRLPGGGRAARRTLTHD QITTMHRKPASMDVTLMRTPVSSLGIRIVGGRAEREATSVVAGGGKPLLEKRFFRLAFDQ LRTDLRVATLNRTPLAQANDAVLELIMHPHVPVHKMYAALFEMEKNHPTLDAIFEEVADW GVKLWNEAAVEGLVQGNITSESATQLVGDVISLLPLKNIVAANTIAKPTISSFSSLRRAS SVAPQSSSKQDSVPSSDMPTSTETLSMPSSAESQTPESTDRASFPMESTTGPAPYSVELS TGRTPYPDDLPTSSTPYPDDLPTSSTPYPDDLPTSSTPYPDDLPTSSTPYPDDLPTSSTP YPDDLPTRSTPAADDLPASSTPYPDDLPTSGTPYPDDLPTSSTPYPDDLPTSSTPYPDDL PTRSTPAADDLPTSRTPYPDDLPIRSTPSPIVLSAGRAPSVVVSPIERTSSPALLPTERT PLTERIAQALRPVGEAAAVRRLLVLRRSSIHRMPYSPFQYSSTWFHPSSNSRRQMMLRMA LRRQLGALTRQAQPDTEGKPSTVFVFRRLAPMPRRLNIEQGPSFLQRQGEYAASGSAPHS DCTNGACAKGACSGTGCQKTPVKPSGGPCKGDACTGSSCNSLECQRSSSSKSDHPCTGDE CPVPCRGPGCSAPASHPSPCTETECGKNSCPGCKQEEEKLVQLRSIRKNLNPNDKKNQAY LLIEVGALPNIHDRAVLYMVSRWMSQRFFNKLRTEQQLGYLTAMHSSRLEDRFYYRFFIT STYDPAEVADRIVEFINAERSKIPTQEEFATLKQAAIDVWKQKPKNIFEEFRKNRRQVVL GDRLFDINERMVAELERVTPEEIQSFKEKTMFNAPWLLMEVYSQREKPQMLPQANGASGE SKLDPWVDIDPELLSNITVPDMESD
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_269885 | 630 S | YYTGSHEDG | 0.994 | unsp | TGME49_269885 | 630 S | YYTGSHEDG | 0.994 | unsp | TGME49_269885 | 630 S | YYTGSHEDG | 0.994 | unsp | TGME49_269885 | 960 S | LRRASSVAP | 0.991 | unsp | TGME49_269885 | 972 S | SKQDSVPSS | 0.993 | unsp | TGME49_269885 | 998 S | QTPESTDRA | 0.99 | unsp | TGME49_269885 | 1229 S | LRRSSIHRM | 0.996 | unsp | TGME49_269885 | 1369 S | QRSSSSKSD | 0.995 | unsp | TGME49_269885 | 1370 S | RSSSSKSDH | 0.997 | unsp | TGME49_269885 | 52 S | RRATSFVDS | 0.996 | unsp | TGME49_269885 | 114 S | VGVGSMSDP | 0.996 | unsp |