

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
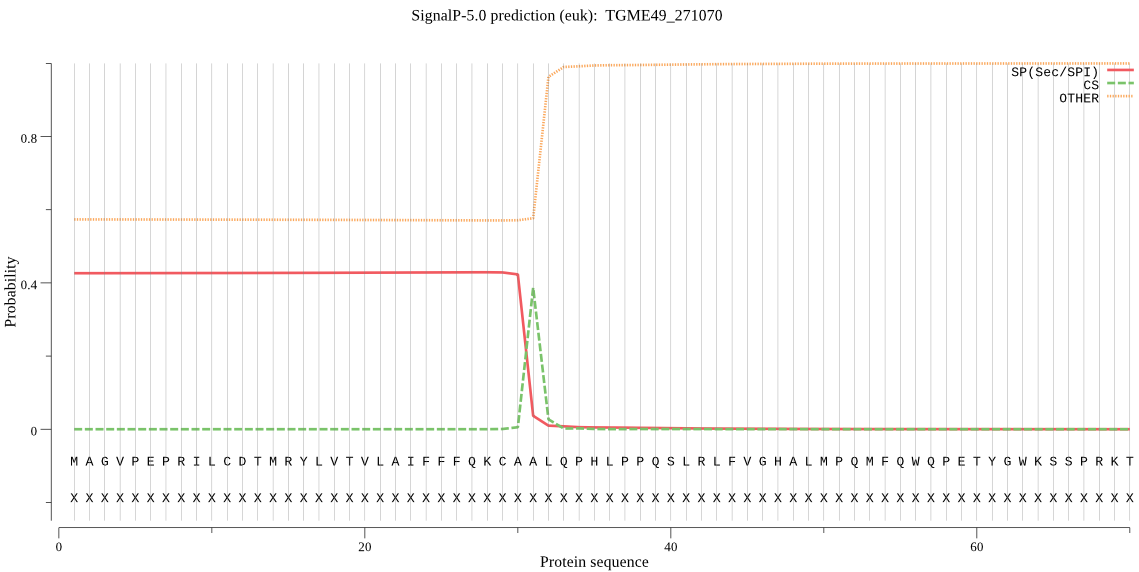
| TGME49_271070 | SP | 0.326469 | 0.658858 | 0.014673 | CS pos: 31-32. CAA-LQ. Pr: 0.9237 |
MAGVPEPRILCDTMRYLVTVLAIFFFQKCAALQPHLPPQSLRLFVGHALMPQMFQWQPET YGWKSSPRKTVRGPHKQNRVFFSGSSSEFDTLQMVRQYSSRDYVSNAVLRRLSRSSQGDT DLDESDNSLLPGCLEVRDMPYCVKAREVPGDGACLFVSVAASLWWNAFETHADLNDPAFV DMVGSLRQLAVDTLQDTNATPLALEGDEVLSRRTLVTMAAADYNMTPAAYCERMRLPGTW GGGPEIVAMSHALRRVIVVYEKHSPSMQSGDSQSGSAFDTHPHRLSRPPATDSPSDSGQN PIKLKVVACLGFPENVAEEPLHILFTSSAGDGQAADHFLPLFPVVSQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_271070 | 113 S | LRRLSRSSQ | 0.995 | unsp | TGME49_271070 | 113 S | LRRLSRSSQ | 0.995 | unsp | TGME49_271070 | 113 S | LRRLSRSSQ | 0.995 | unsp | TGME49_271070 | 116 S | LSRSSQGDT | 0.998 | unsp | TGME49_271070 | 264 S | YEKHSPSMQ | 0.993 | unsp | TGME49_271070 | 272 S | QSGDSQSGS | 0.991 | unsp | TGME49_271070 | 293 S | PATDSPSDS | 0.996 | unsp | TGME49_271070 | 87 S | SGSSSEFDT | 0.996 | unsp | TGME49_271070 | 99 S | VRQYSSRDY | 0.997 | unsp |