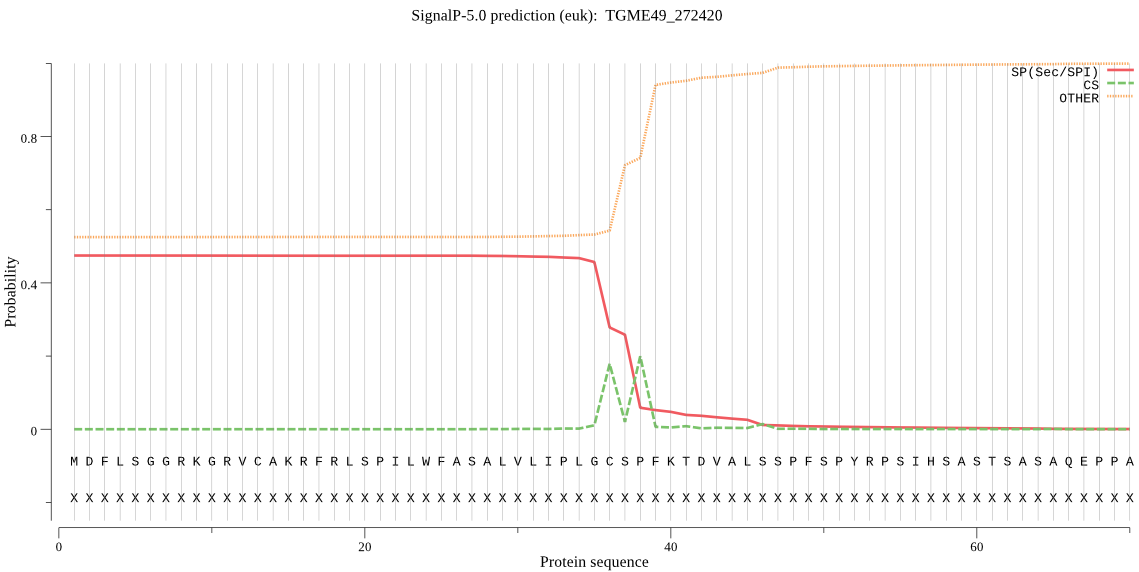
Fasta :-
MDFLSGGRKGRVCAKRFRLSPILWFASALVLIPLGCSPFKTDVALSSPFSPYRPSIHSAS
TSASAQEPPASVHPTPSAVPSDPQVASRVGPHGATKSGFSRATRENVPAFPSRDPSFSTS
VPSSTVSETDSGIFVVSSQDPVANQDHGISSVPSTASEPDSGVDHALFVIPGIAGSGLFA
TVTNASVASCGKSPISYPNPFRVWASLSLLLPPATHQKCWIDMMKMVVDENGEVYTGQEG
VRVEVDGYGGIHAIDYLDYYMNNTYGVPASAYMHVMLRTLMSLHYSQFVTLRGVPYDWRL
PPWQLDYAQLKTDIEDRYTEMNNRKVDLIAHSLGSIILCYFLNRIVDQAWKDKYIGSMTI
VAAATGGSFKAIKSLLSGYDDATDVDIWNVIDISLFPAGLLRDLLQTMGSIFALLPDPAI
YGPDHVVARVARPTQVPASSSASAFAPLTGRSDRAAKKAEGTTPTALRWSHSGEMRRLDA
VLTDEVGGPESGARELKSEQAFSLLSELAKDNVEVEKEEAEGETNTRFHKYPDFAPGDQM
EEEELAKAEERRDALLREFQLQNRLSHEDRAARQRERYSLFVEKEQRRLEEALRVAQEQR
LEEDVYTLSNWTSLLPANLQRRVKTAQEKMAGVVADPGVPVRCIWSKFSQPTTDVAYYYP
TGSLDTHPIRIFDYGDNTVPLASLSLCASWPSTVQTKVFDNLDHMFLFADRGFNNFIFDI
FSPDANPFLSSLLGEQEEVMKRDIPSPQESVPAGRGAIMADST