

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_272510 | SP | 0.004799 | 0.991058 | 0.004144 | CS pos: 21-22. STA-AK. Pr: 0.4462 |
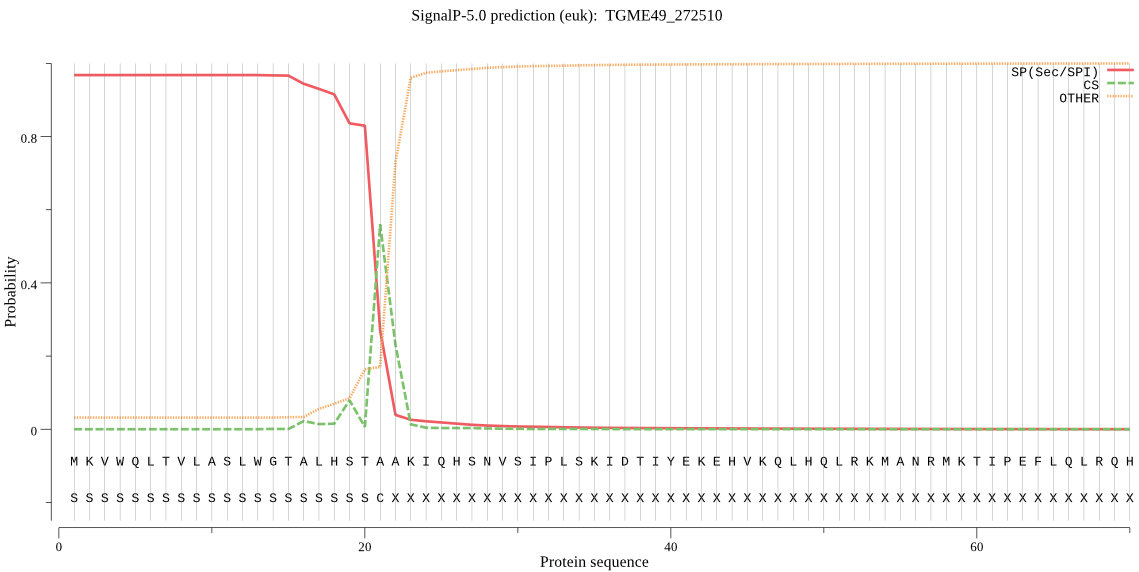
MKVWQLTVLASLWGTALHSTAAKIQHSNVSIPLSKIDTIYEKEHVKQLHQLRKMANRMKT IPEFLQLRQHPLSSVSEIPLHDQVIASKKLATYYGEISIGSEPERAFKVLFDTGSCEFWV PDETCLSMQCIGHTKYRRSASFQPRFNQNGKPSLMNVVYLSGTLQGYDGYDTVHLGNGIS VPHTNIGFGTLVDIPLLRDLAWDGIVGLGFKNHEISSRGVVPFLDHIVQTKALESKHLGN QFAYYLNADGGSMTFGGADLSKKEYPTEEFSWIPVDPSDSYWTVKVIGVHKEPRSTSSLA TVTKQLENGQTAVTQTDGKKSIVDTGTYLIYAPATVMENELSDISVSSCDDKKRLPDLIL QFLGNPQHGGDSVVEVRLSPDDYVLEFLEDDGSRECMLGIAMDDAAEEDALDGWTLGQVF LRCYYTVFDRDNLQIGFARAKH
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_272510 | 261 S | GADLSKKEY | 0.995 | unsp | TGME49_272510 | 261 S | GADLSKKEY | 0.995 | unsp | TGME49_272510 | 261 S | GADLSKKEY | 0.995 | unsp | TGME49_272510 | 321 S | DGKKSIVDT | 0.997 | unsp | TGME49_272510 | 347 S | DISVSSCDD | 0.994 | unsp | TGME49_272510 | 379 S | EVRLSPDDY | 0.998 | unsp | TGME49_272510 | 34 S | SIPLSKIDT | 0.993 | unsp | TGME49_272510 | 139 S | KYRRSASFQ | 0.99 | unsp |