

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| TGME49_272670 | OTHER | 0.862151 | 0.000496 | 0.137353 |
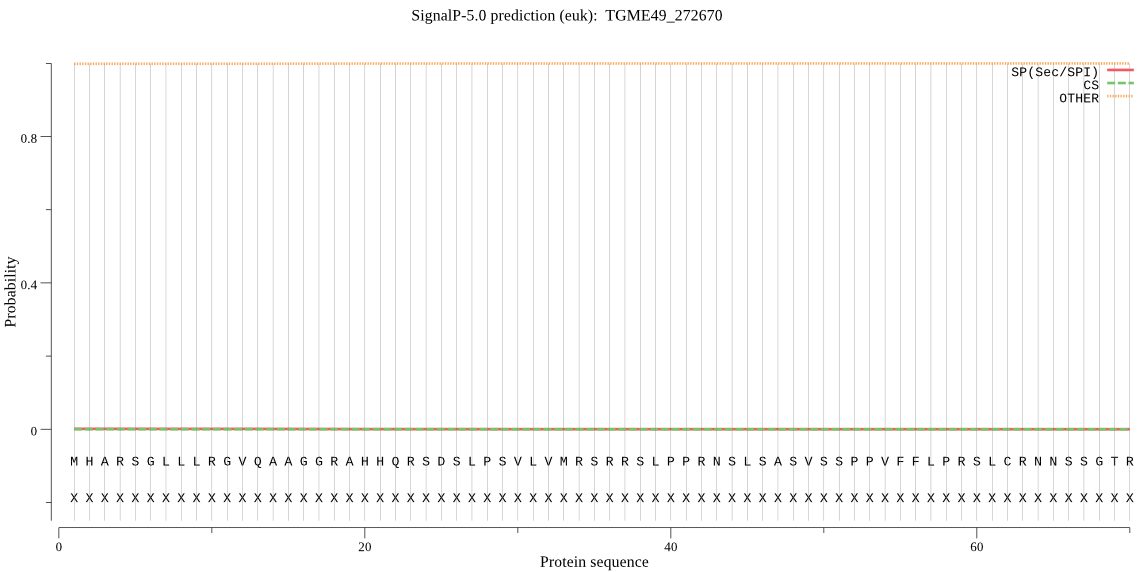
MHARSGLLLRGVQAAGGRAHHQRSDSLPSVLVMRSRRSLPPRNSLSASVSSPPVFFLPRS LCRNNSSGTRSSHSSSLTFHRVPPASSTLERRTLRRPFASLSRLSHSGQSEWRRQPHGLC SSPPCFSSTFPTPSPPRKPGLFSFLSGLTQISSRGSRAAGSDELRPGLLQLPGVHTPEQL VRLGRQAVADCEAFLATGGAPTSSSSGSREEGKLSSPSSCPSSASSGDASDVSSSWKEAR RAVQALDAVSNTLCKVADGSELLRQVHPDPRWKQAGAEVVAHVQDFMSRVNVNNKVYDKL HRLTEALGDSLSREAQAPSCLSLPSSSTSASSSPSTSYPSSSSLPSSLSSSLSASPSSAS SEDEGNMAEGDEGAGVSSAQTENAEKHRLTTEEILVLRSMRDAMEQQGVFLEPQVKDRYL RLLTEEATLAAGIAERQETPQSTEGFLVSTDVLLASGVPASLLAQLQGEKRSFFSPRRRP LCRRGGSGGDQVLVAAHSELAHVILRQSADSGLRREVYELQRLPGKDDKETEKQLLRLLH LRQELAKLRGYPTWTAYAQRDGILKTPDKINAFLSRLLDALRPGVESDLKTLSVFGASSG LLRDAKGLDPWDLPFLLRAYSVRTPDGSTGARSISIRDLLEGMQSVVERLLGLRFVRAPT AKGETWHWSVLRYELHEAPSSRASRLFSFSPSSSASPPSSSLDASSSSLRGVLYLDLWAR PSKTRLLAQFTVRGSKRLSYAHGQGWRLGGPLWLSAVQRLSREGAASEDEASATATVAVE EELRQIPCAALVCSLPPPPEVDMTQPTGPDVDRLLEATSLSASLTRSLLHECGHVLHSLL SETELQHLSGTRGAVDFAEFPSNLFEHFSAPLLDGYLASPTRRARPALGSDGRGKACNFA HLGALQLLLHACVDQAFYSFVPGVSELPPCSGDARAPSPPPEAVDAQLAALRGCLDTAFS RHHWLAGDPRPGVTEQPICDVVGRPQMARFEHLVHYGGSYFCYLLCRVMSSFAWQHAFAK DPFDRETGKRLHAILRRGSIDCSLRPILELAGPGVLSETQKEQLLNNPHLLPVEPFLEEL HADDASAAAELPFP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_272670 | 105 S | LSRLSHSGQ | 0.996 | unsp | TGME49_272670 | 105 S | LSRLSHSGQ | 0.996 | unsp | TGME49_272670 | 105 S | LSRLSHSGQ | 0.996 | unsp | TGME49_272670 | 156 S | SSRGSRAAG | 0.992 | unsp | TGME49_272670 | 206 S | TSSSSGSRE | 0.994 | unsp | TGME49_272670 | 208 S | SSSGSREEG | 0.998 | unsp | TGME49_272670 | 226 S | SSASSGDAS | 0.99 | unsp | TGME49_272670 | 235 S | DVSSSWKEA | 0.998 | unsp | TGME49_272670 | 333 S | SASSSPSTS | 0.993 | unsp | TGME49_272670 | 355 S | SLSASPSSA | 0.991 | unsp | TGME49_272670 | 360 S | PSSASSEDE | 0.997 | unsp | TGME49_272670 | 361 S | SSASSEDEG | 0.993 | unsp | TGME49_272670 | 399 S | LVLRSMRDA | 0.996 | unsp | TGME49_272670 | 475 S | RSFFSPRRR | 0.993 | unsp | TGME49_272670 | 487 S | RRGGSGGDQ | 0.996 | unsp | TGME49_272670 | 511 S | QSADSGLRR | 0.991 | unsp | TGME49_272670 | 635 S | ARSISIRDL | 0.997 | unsp | TGME49_272670 | 700 S | SPPSSSLDA | 0.991 | unsp | TGME49_272670 | 761 S | VQRLSREGA | 0.996 | unsp | TGME49_272670 | 767 S | EGAASEDEA | 0.996 | unsp | TGME49_272670 | 1039 S | LRRGSIDCS | 0.995 | unsp | TGME49_272670 | 72 S | GTRSSHSSS | 0.992 | unsp | TGME49_272670 | 74 S | RSSHSSSLT | 0.993 | unsp |