

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
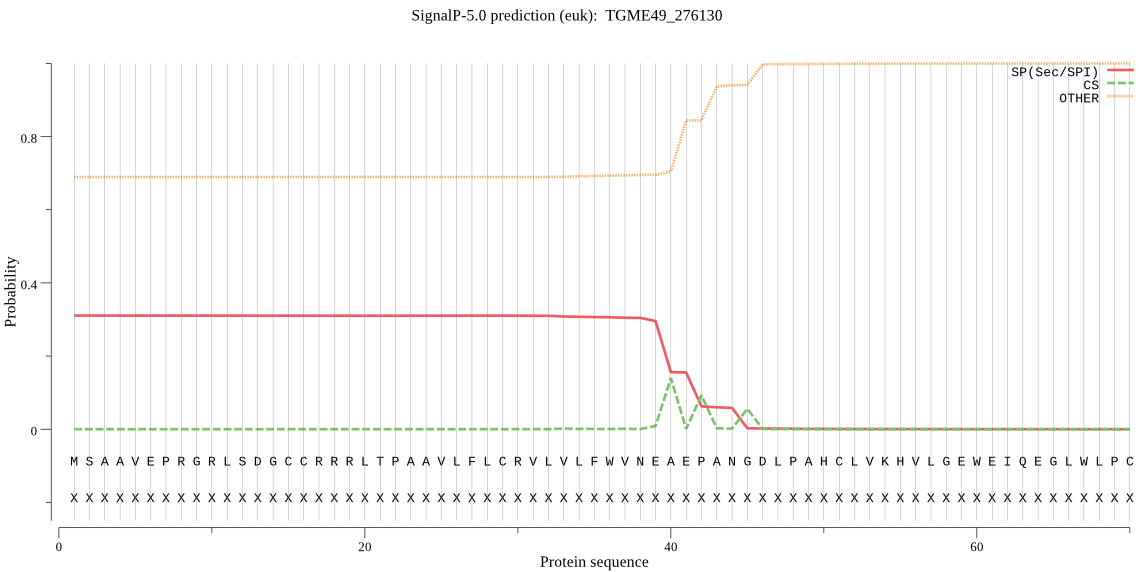
| TGME49_276130 | SP | 0.386514 | 0.613146 | 0.000340 | CS pos: 45-46. ANG-DL. Pr: 0.3419 |
MSAAVEPRGRLSDGCCRRRLTPAAVLFLCRVLVLFWVNEAEPANGDLPAHCLVKHVLGEW EIQEGLWLPCQEEGNLTTDAARLHDPYCGYGIPDRVDAHGPMTPPNVSASFHALRTSRFT LFPDFRIEFEDGSSGLWTLVYDEGLHFEVSTPQNRRFFAFFKYQLAPDGQNRAWSYCHTT LVGWWERPAAPNLSLQDVEKAPPVYDIPEPIAPDTAADALLKSVKRGCWWGRKVGEDLSV PTNEVPKERRTPLEVPLWPTSRPPDVRTVAAVVRKNLAADAPWEPLDEEYVEIRGRKLRT MEEVWAHAGHQPLLISRQNIESTHLTLSVRPPLHLLAGIKPFGSPAGSDPPWRTVRDFDW ANEDHVALRIGRRQSVIPDAPNQGGCGSCYAIATGTVLTSRLWIRYAANDDVFGKVNVSA FQGTSCNVYNQGCGGGYVFLALKFGQEHGFRTEDCVSEYHAMADKHKGSLSPNLQTCFDL GGQLGTSAFGCQAPPARASLPDSCNLSVKVTSWHYVGGVYGGCSEDDMLRTLWEHGPMAA SIEPTIAFTVYKKGVFRAAYNSLVEQGDNWVWEKVDHAVVISGWGWAKHGDSWLPYWKVR NSWGTKWGEGGYARVLRGVNEMAIERVAVVGEVSLFRGGEHIPPLLARSLDSQGSSNASK DSERGETLVESVSPVFRGSASGSESGNPRDAATNESADMSESADADAESSSGSDDDEDGD MEKGDVESTTKQTTLFRPIQRHGEVLEEKRTGE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_276130 | 375 S | GRRQSVIPD | 0.995 | unsp | TGME49_276130 | 375 S | GRRQSVIPD | 0.995 | unsp | TGME49_276130 | 375 S | GRRQSVIPD | 0.995 | unsp | TGME49_276130 | 499 S | PARASLPDS | 0.994 | unsp | TGME49_276130 | 681 S | RGSASGSES | 0.99 | unsp | TGME49_276130 | 711 S | AESSSGSDD | 0.997 | unsp | TGME49_276130 | 713 S | SSSGSDDDE | 0.997 | unsp | TGME49_276130 | 21 T | RRRLTPAAV | 0.992 | unsp | TGME49_276130 | 223 S | ALLKSVKRG | 0.991 | unsp |