

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
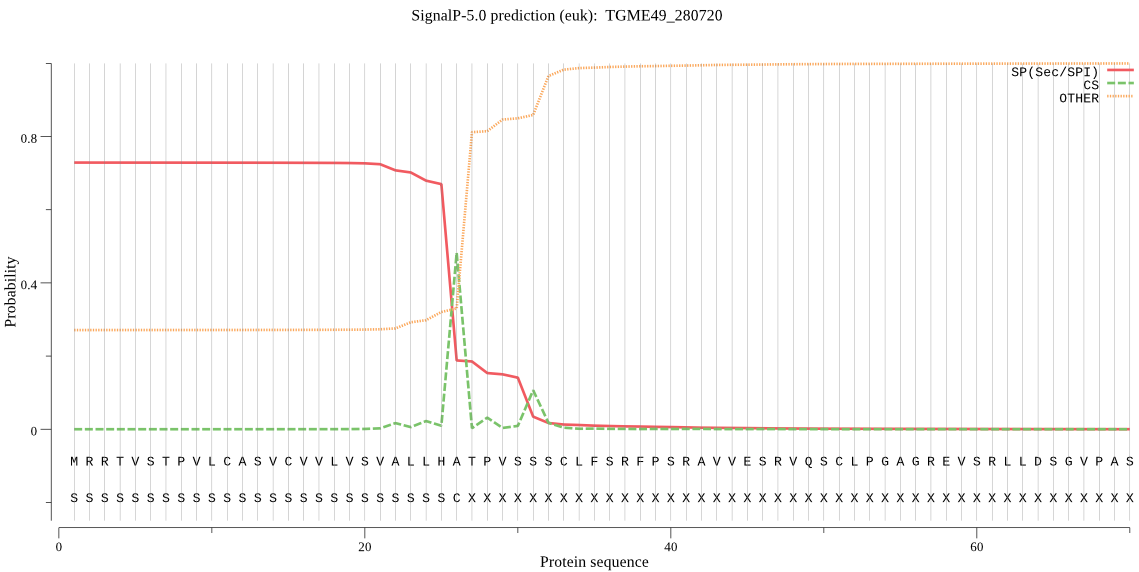
| TGME49_280720 | SP | 0.465091 | 0.530665 | 0.004244 | CS pos: 26-27. LHA-TP. Pr: 0.5331 |
MRRTVSTPVLCASVCVVLVSVALLHATPVSSSCLFSRFPSRAVVESRVQSCLPGAGREVS RLLDSGVPASAPLRRLQPRDARLSRPFLTLPSSSSSSLLSSSLSPRVSSSRNRLLPAAWL SVLRGCMRAVSRTRTSEGQPFLLPLPFSTFLRLHFLTTLLSFTALGFNAFLSSRRLVLLP SLPRACLQLLNGNNQLHVFLRSVCTPDLPALQATVSGPFFEELQFRFLLQNCLLSPLLAA VGSSWLSVAAGPGCRRLSPVKLEDGETTVRRDGQGASSPSDATASSKENAESTHPEARKR RLGSVEGSSLREKLARVLRCARLVETRERFFRDVSGANSSEKDLRAGCQQQLLQERREGS SPRCRVEQTLRIAITSILFALAHYVPPSVASPVRRQFRRAVAKQRTRLGFPLEPSGAKRF SIGSVSSFSSPSVKQWSPRSAAQSAAPDAASFSSSTAFTSVPLAEEELFSVDSPQRRVED CVAANRILTAAVQGCVWSYAAEKGGFASALLLHILHNLQQFLLVAAFRTFVRRRQERLSD GRLKESRVRERISSRP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_280720 | 136 S | RTRTSEGQP | 0.995 | unsp | TGME49_280720 | 136 S | RTRTSEGQP | 0.995 | unsp | TGME49_280720 | 136 S | RTRTSEGQP | 0.995 | unsp | TGME49_280720 | 258 S | CRRLSPVKL | 0.997 | unsp | TGME49_280720 | 268 T | DGETTVRRD | 0.992 | unsp | TGME49_280720 | 278 S | QGASSPSDA | 0.998 | unsp | TGME49_280720 | 285 S | DATASSKEN | 0.995 | unsp | TGME49_280720 | 286 S | ATASSKENA | 0.994 | unsp | TGME49_280720 | 304 S | RRLGSVEGS | 0.996 | unsp | TGME49_280720 | 309 S | VEGSSLREK | 0.99 | unsp | TGME49_280720 | 340 S | GANSSEKDL | 0.997 | unsp | TGME49_280720 | 361 S | REGSSPRCR | 0.995 | unsp | TGME49_280720 | 437 S | VKQWSPRSA | 0.993 | unsp | TGME49_280720 | 539 S | QERLSDGRL | 0.997 | unsp | TGME49_280720 | 104 S | SSSLSPRVS | 0.991 | unsp | TGME49_280720 | 108 S | SPRVSSSRN | 0.995 | unsp |