

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
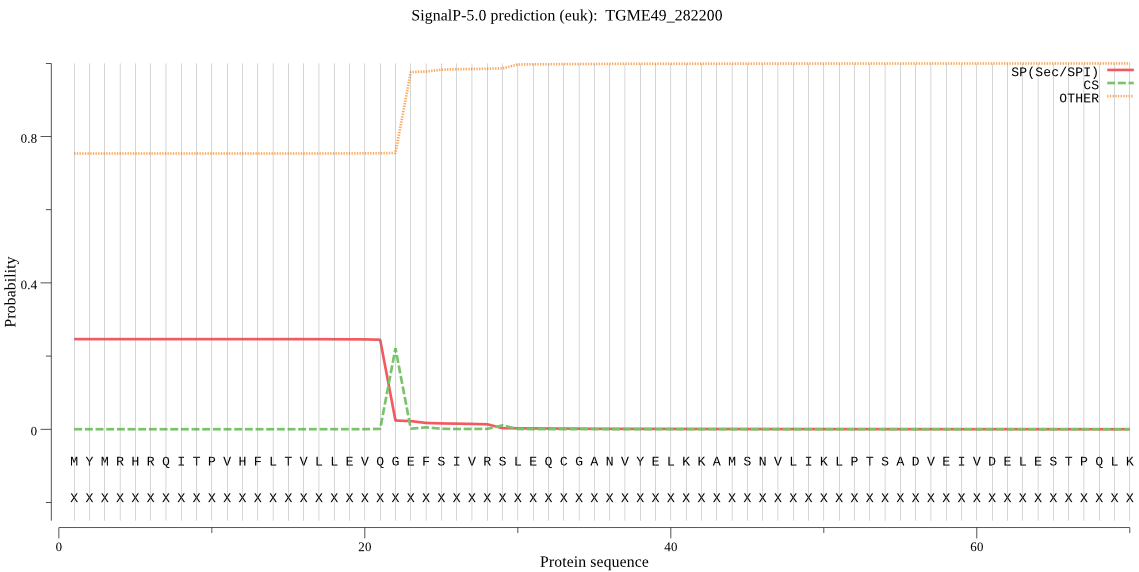
| TGME49_282200 | OTHER | 0.908788 | 0.079458 | 0.011753 |
MYMRHRQITPVHFLTVLLEVQGEFSIVRSLEQCGANVYELKKAMSNVLIKLPTSADVEIV DELESTPQLKRVALRALEIAQKDKKQTIGMEHLITSLFEENNLRKSLEQAGCNVKVFLEH STTMYENKFKQDVASRSMKKQPTEGSPATSRAKPDASDEEFVQSFGVDMTKLAAEGKLEP VVGRNKEIKEVLTVLSRKGKGNPCLVGEPGVGKTAVVEGLAQRLVEGMVPKSLENKILFA VDLGALIAGATYRGEFEKRMKALIRYAVNQEGRVILFIDELHMLMGAGKSDGTMDAANLL KPPMARGEIRLVGATTQEEYKIIEKDAAMERRLKPIFIEEPSTDRAIYILRKLSDKFESH HEMKISDEAIVAAVMLSHKYIRNRKLPDKAIDLLDEAAATKRVKWDLRSLNDDEVAMERK NSEQFEEETRKVEQQDQQRIEKQNHGDGTEEELKNLEKEIEEQSLHEKQELVLTADDVAQ VISLWTGIPLAKLTDDEKSKILRLSDLLHSRVIGQDDAVKAVADAMVRARAGLSREGMPV GSFLFLGPTGVGKTELAKALAMEMFHSEKNLIRIDMSEFSEAHSVSRLIGSPPGYVGHDA GGQLTEAVRRRPHSVVLFDEIEKGHQQILNIMLQMLDEGRLTDGKGLLVDFTNCVIILTS NVGAQYIISAYEQAEKESKTALATFDPKSGNLEQFAEAAKGAEAASEGDDSSSEKKDKKG AVLDKNKKSGNWKKEARRKILSEIAGSGLLKPEVVNRFSGVIIFEPMNPADVRKILAIQS KDLKNALQRKGIELVIDASAEEFIVQRSYTHKFGARPLKRFVDNHIGAKMAPWILSGYLQ PGMRVTVVASKRNKNTLEAHVCKVVMGKCDRATRKARVLASVAKKDDSSPDDPGR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_282200 | 157 S | KPDASDEEF | 0.996 | unsp | TGME49_282200 | 157 S | KPDASDEEF | 0.996 | unsp | TGME49_282200 | 157 S | KPDASDEEF | 0.996 | unsp | TGME49_282200 | 354 S | LRKLSDKFE | 0.996 | unsp | TGME49_282200 | 584 S | SEAHSVSRL | 0.992 | unsp | TGME49_282200 | 614 S | RRPHSVVLF | 0.994 | unsp | TGME49_282200 | 706 S | AEAASEGDD | 0.995 | unsp | TGME49_282200 | 711 S | EGDDSSSEK | 0.995 | unsp | TGME49_282200 | 713 S | DDSSSEKKD | 0.996 | unsp | TGME49_282200 | 888 S | KKDDSSPDD | 0.997 | unsp | TGME49_282200 | 889 S | KDDSSPDDP | 0.998 | unsp | TGME49_282200 | 106 S | NLRKSLEQA | 0.993 | unsp | TGME49_282200 | 137 S | VASRSMKKQ | 0.994 | unsp |