

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
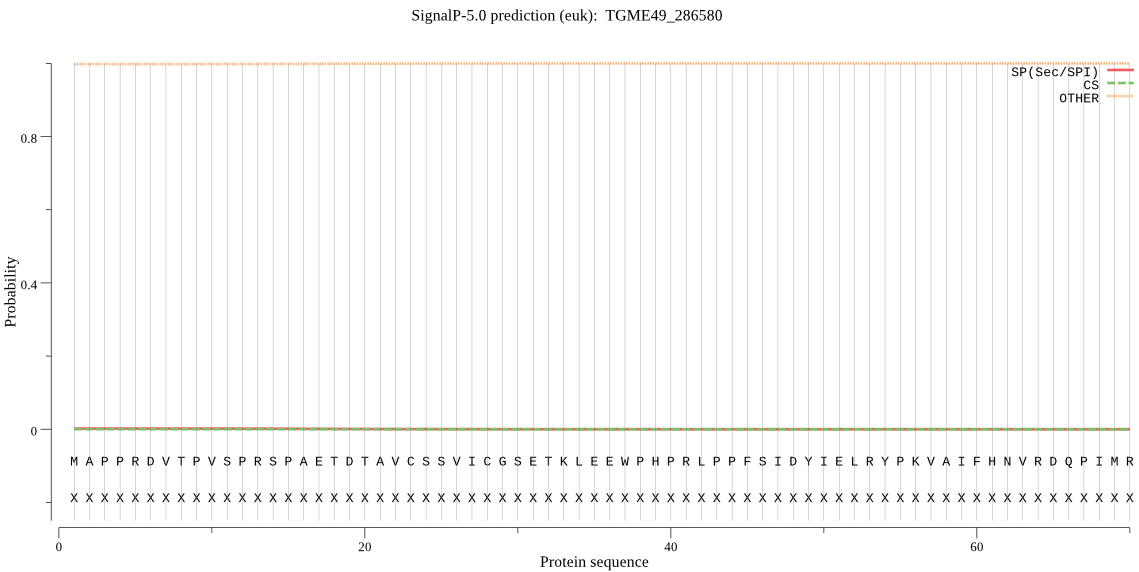
| TGME49_286580 | OTHER | 0.999947 | 0.000034 | 0.000018 |
MAPPRDVTPVSPRSPAETDTAVCSSVICGSETKLEEWPHPRLPPFSIDYIELRYPKVAIF HNVRDQPIMRYTWTTTTGPARGAVVLLHDQGTHTMFEWLKHLPPLSIEERQQMEEDTEAT EVEKKEFMRLRKASHYEDSWVQKFNDAGLDVHAIDLQGHGLSKVVEGKNYELPSSFDEFA EDACWLLEEVCRNSTLPIFIMGQGLGATIAARAVEMLARQGKLGKPHVPPAKPADAVGAS EQSTTTGTSQSSEAGREVPGMQYATDPQYCLHRGKGKKEVPVEHITVQARPIPIAGLVLL SPLLTLKEITPEEMRKGKGKHTEGIMTRIARCFGSTRDSSHPLGGLMGFRMHQQNPLMPE YNVWYKNDTMTIKGLTPQTMYTQIAHGIDSAMDDARWLLTPVERDSTQTFSLKDWQEKHR RWMEMMRTKRPNVLATVVGENKKNASEAPDLAPWLKDEVENQDSSVKPWRLHELAAAQEA EKNASRLPQERILSERAKLAYYVNVLLVQSLADTTANPRGAAYFFKRIGGRIQGSRDILV QLGELPEEQGHSGSCVSCVHPDACQEDESEKDTAVEFSTENDDSGYVEVVTNRPLSAELS VQSQTSQPRPVLTHTGSRSFGETQDSRYEDTVEGLCLGSVTSHSGESAHENSEARDEAAS HEDAQSQETAQGDNQGVHERVKKLMEVNKILDKEMQLATMEEGTDGILRPGTFVRPSHLR RFDTEDIQRAKREAAEGFVGVEEVQWVKEVPASGAGLLACCGGSTGRVQRVDLVEAKVRL VDKIVMMEATDTVKAMIQEIEERERLQNEKANERRQLGGWFFNCTAEEKPAEKPVPEPVI SQQKRLWRQVQGVIVDESGRRVPVEEVERLRTQKPEDYMRLFFRYGAATSSGNHARTAMW LVPDMYHSLCQEDKEGKLVSRLLDRWILPVLGDLEERELVWQEMNRSAKDVEAAQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_286580 | 134 S | LRKASHYED | 0.997 | unsp | TGME49_286580 | 134 S | LRKASHYED | 0.997 | unsp | TGME49_286580 | 134 S | LRKASHYED | 0.997 | unsp | TGME49_286580 | 411 S | TQTFSLKDW | 0.992 | unsp | TGME49_286580 | 465 S | NQDSSVKPW | 0.99 | unsp | TGME49_286580 | 569 S | QEDESEKDT | 0.998 | unsp | TGME49_286580 | 578 S | AVEFSTEND | 0.995 | unsp | TGME49_286580 | 596 S | NRPLSAELS | 0.997 | unsp | TGME49_286580 | 626 S | ETQDSRYED | 0.994 | unsp | TGME49_286580 | 644 S | VTSHSGESA | 0.994 | unsp | TGME49_286580 | 647 S | HSGESAHEN | 0.991 | unsp | TGME49_286580 | 660 S | DEAASHEDA | 0.998 | unsp | TGME49_286580 | 11 S | VTPVSPRSP | 0.995 | unsp | TGME49_286580 | 14 S | VSPRSPAET | 0.998 | unsp |