

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
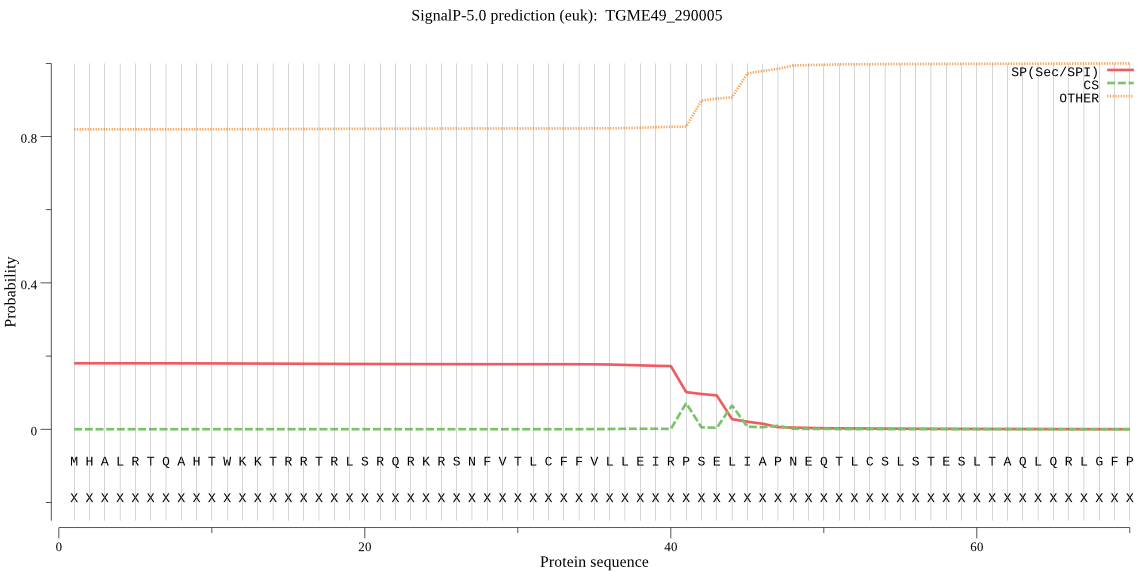
| TGME49_290005 | OTHER | 0.978388 | 0.008696 | 0.012915 |
MHALRTQAHTWKKTRRTRLSRQRKRSNFVTLCFFVLLEIRPSELIAPNEQTLCSLSTESL TAQLQRLGFPNFLSVFVLVVFFAFSITFSSVFFSSVFSFSPFSRFFSGAFNDSIMLAGGP ASLPTSRLLHPEEEAILPVALAAPMERRFSPYVNNGGTVVCVAGEDFAVAVGDTRLSTGF SIYSRRQSKITKLTDKVVLATSGMEADKTTLHNLLKIRIEQYTHQHRHPPSLNAIAQLLS TVLYSRRFFPFYTFNVLCGIDENGKGAVYGYDAIGSFEPSRYNCAGTGAHLIMPVLDNQI SRNNQQGEKAPLTRERIVEIVKSAVASAGERDIFTGDQAEVVMISQSGIEKTLMDLRAD
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_290005 | 20 S | RTRLSRQRK | 0.995 | unsp | TGME49_290005 | 150 S | ERRFSPYVN | 0.997 | unsp |