

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
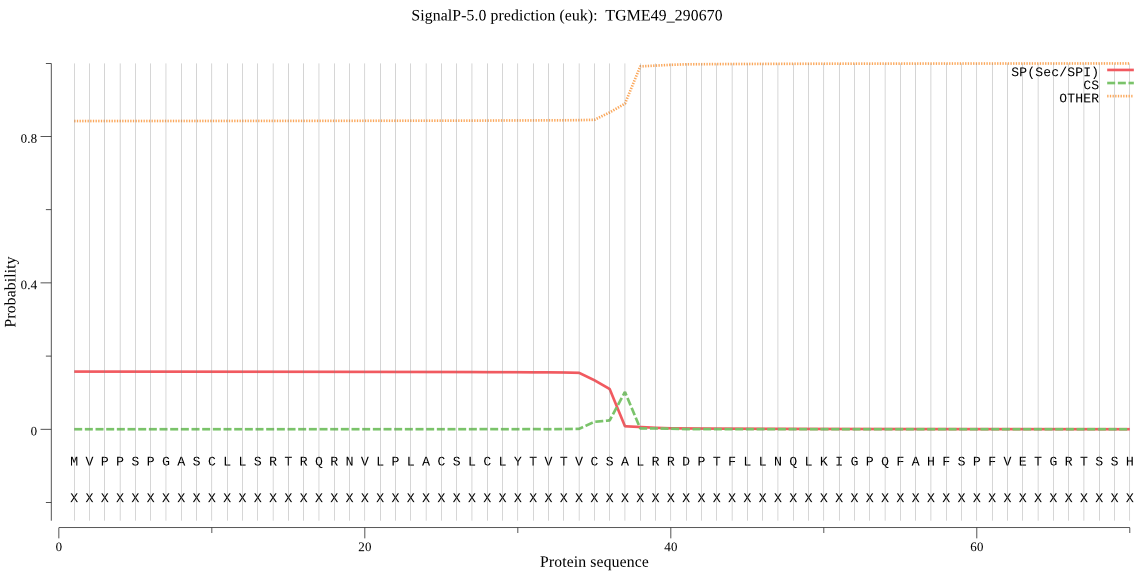
| TGME49_290670 | OTHER | 0.874353 | 0.119611 | 0.006036 |
MVPPSPGASCLLSRTRQRNVLPLACSLCLYTVTVCSALRRDPTFLLNQLKIGPQFAHFSP FVETGRTSSHSGVSPSTFSGSLRSRQHAFTMRPSSFPAFLVKLPAKHKNQCRQSAAAVAG STACWQSSLHAPACAFPFSRRNGGHPERRDAPSLSSPRSSPASFLSVLRPVTSPLPCVWK AKTPHARPLALPLPPVRTASLATFSSGSSPRRSSNHSEMSRVPAPLTVVKTDPTAIPFVG CQPAKKVRMEAVDISEAEKYSGDLIVFSVLAPGCFHDAPNASQPQKGPIALPELAQKFDD AVGQGMLKDAVEAADFKAKMGSQVFVRLPSKSAPAVKQLAALGFGSIAEVTPSAVTKAAA AFASMLLRGSGEKAKTVAVVLPTCQKVCPSAEGQRKTREMKERVVTSFLETLLVELQPDL RFKGDREDGKHSGPQVEKLTLFTDDVEAVNRAIARAKVVAPGVYFAQELVNAPANYCNPV TLARAAVEMAKEAGLEAEVLQQDDIEKLKMGCYLAVAKGSLFPPQFIHLTYKSSNPKRRV AFVGKGICFDAGGYNLKTGGAQIELMKFDMGGAAAVLGAARALGELKPENVEVHFLVAAC ENMISAKSYRPGDVITASNGKTVEVGNTDAEGRLTLADALVYAEKTVKADTIVDVATLTG AVIVALGYKYAGLWSNKECLASSILKSAENSGECMWHMPLAKDYAEALDSKCADMNNVGG KGKGGSIIAAVFLNKFIEQASWAHIDIAGTAWDMKTSMATGFGVRTLVEYIMDISTQTKE N
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_290670 | 156 S | PSLSSPRSS | 0.993 | unsp | TGME49_290670 | 156 S | PSLSSPRSS | 0.993 | unsp | TGME49_290670 | 156 S | PSLSSPRSS | 0.993 | unsp | TGME49_290670 | 160 S | SPRSSPASF | 0.994 | unsp | TGME49_290670 | 209 S | SSGSSPRRS | 0.997 | unsp | TGME49_290670 | 213 S | SPRRSSNHS | 0.992 | unsp | TGME49_290670 | 214 S | PRRSSNHSE | 0.994 | unsp | TGME49_290670 | 370 S | LLRGSGEKA | 0.995 | unsp | TGME49_290670 | 608 S | ISAKSYRPG | 0.996 | unsp | TGME49_290670 | 68 S | TGRTSSHSG | 0.994 | unsp | TGME49_290670 | 71 S | TSSHSGVSP | 0.991 | unsp |