

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
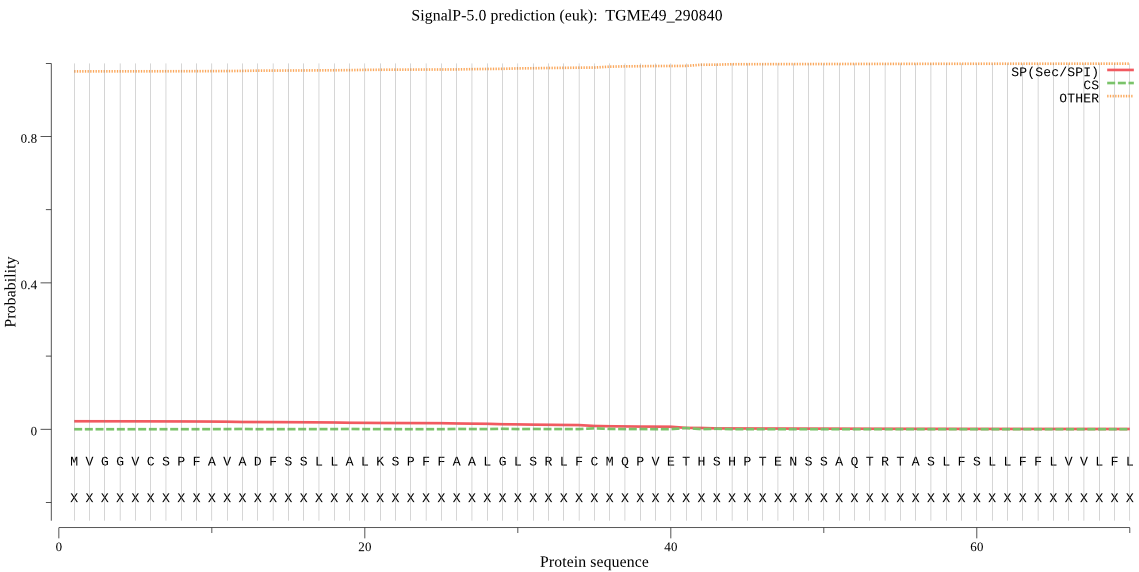
| TGME49_290840 | OTHER | 0.996662 | 0.000577 | 0.002761 |
MVGGVCSPFAVADFSSLLALKSPFFAALGLSRLFCMQPVETHSHPTENSSAQTRTASLFS LLFFLVVLFLVRSLVCPRFFFSVAFLSSRFNATTVLVAPLFPFLASLAHPAPSFGIASSP SLLSSVALSLHPSPLASSPRSSPLLPSHPSPPSSRLRLPASSSLSSAFRRSPRGCPSAVR LCRLYAHREGSSRGSPVSASGASQPRSLACTMASAVLLRRLGFVGSPSASVASLQGDLAA RHTTQRLLSLAFARRVNSPRSPNQAKRLFSSSSLFSPSSSSSLFSSTLASRCRSACQSSS PPSFLADRRRLSSSLSASLSSLSSLSPALRPSNGGDPASPRRSSPASLSRSAAFSSASFP VPSQLEGDVLSEPPSGVCTPDGVLGEGAFAQHAAPGVADALFHGEMPSPASLSFLFNSVV KVYSDFTDPNYSLPWQMQRQGTSTGSGFVLRDRLIMTNAHCVSWNNRLQVRKHGSPNKFV ARIVAVGHECDLALITVDDEAFWQGDLAQLEFGDVPALQDAVVVLGYPRGGDNLCITSGV VSRVDVNPYAHSNTCLLCVQIDAAINPGNSGGPALKDGRVVGVAFQGFDNAQNIGYIVPT TVIKHFLDDVKRHKGVYTGFPSAGIVFQHLENKSMQAFLGLDKIQPRQLPPGVEASGILV TMADELRARQFARHLEGGAQSAGKREDGERGDSTETNQAGKKKEEPASDSDEDVRLKDGT RVGLKKNDVILAIDGVDVANDGTVFFREMERVNVSHTISSKFIGDTLRATVLRKKEVVDV LVPLIEENALVPKHQWDKKARYLIYGGLVFCPLTLEYLKDEFGTKFSERAPASLLQPLAD IFAKEEGEEPVILSHILASDLTSGYTFRNCLLTHVDGQKVLNMKHLASLLGLPLPASSPS SAPSSVSSSPAESSEKKENDFVIFLLENKVQLVLERSKAESMQPFILKQHAIHSPTSEVL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_290840 | 191 S | HREGSSRGS | 0.992 | unsp | TGME49_290840 | 191 S | HREGSSRGS | 0.992 | unsp | TGME49_290840 | 191 S | HREGSSRGS | 0.992 | unsp | TGME49_290840 | 195 S | SSRGSPVSA | 0.997 | unsp | TGME49_290840 | 258 S | RRVNSPRSP | 0.994 | unsp | TGME49_290840 | 276 S | SSLFSPSSS | 0.993 | unsp | TGME49_290840 | 312 S | RRRLSSSLS | 0.995 | unsp | TGME49_290840 | 339 S | GDPASPRRS | 0.996 | unsp | TGME49_290840 | 344 S | PRRSSPASL | 0.997 | unsp | TGME49_290840 | 347 S | SSPASLSRS | 0.994 | unsp | TGME49_290840 | 693 S | ERGDSTETN | 0.995 | unsp | TGME49_290840 | 708 S | EEPASDSDE | 0.998 | unsp | TGME49_290840 | 710 S | PASDSDEDV | 0.996 | unsp | TGME49_290840 | 905 S | SAPSSVSSS | 0.991 | unsp | TGME49_290840 | 909 S | SVSSSPAES | 0.997 | unsp | TGME49_290840 | 913 S | SPAESSEKK | 0.993 | unsp | TGME49_290840 | 138 S | PLASSPRSS | 0.995 | unsp | TGME49_290840 | 171 S | AFRRSPRGC | 0.996 | unsp |