

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
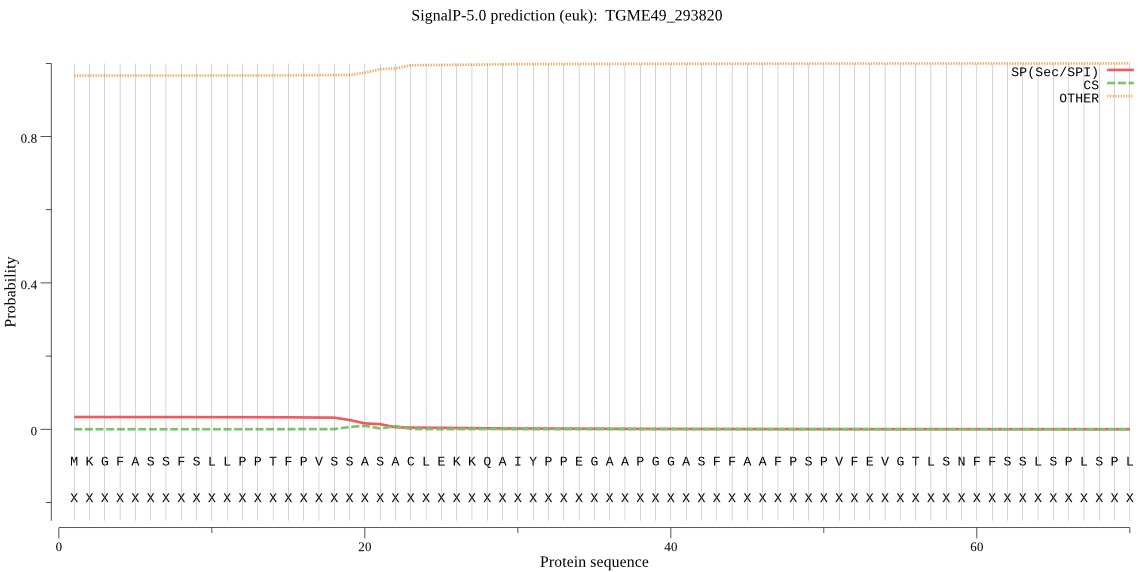
| TGME49_293820 | OTHER | 0.913325 | 0.086620 | 0.000056 |
MKGFASSFSLLPPTFPVSSASACLEKKQAIYPPEGAAPGGASFFAAFPSPVFEVGTLSNF FSSLSPLSPLTSGDHFYSSSHSSPRSTPSSGNAVPSLHSVPPAHAPSGSSQCTDSLPTAR GSCSSLQTKVPDTPQRRRCSLWTAAPSHVWEERWRVDCQAGWERAIEESLDLRSEKEDTA LYREQTPPCIETAVSAGCEGHRRSNSSSPRGASPSVASDRPLASLCVGEELSRAAASPIR APEASCSPSSICPSALPSTREAGPLSPGVAREFSPSVARLRRHSRFRLQDASGSQKGAAE ARDARKLRGFFGTFLADGSGDTRVDPERDPSKPPRHLETRLFNPALRFLSERRIQASSSS CSVFLSSPSPFRGNYLERGVPDCGRDSSPAVVAFGTAPYTRRLSPWSPLRSPNSASACAQ PPRRRNGIAASSARAPSFSPSLSSHSSSLSSYRSSVPSLSPSHVVSSRLSLSSVSSAFAR PFRRVLRRPETWSPLPSEGCPAWRLSPATPEDSLPLSGSLSAASAPSGVRLTASALSPER RPDSLGIASAPPNCLRKMGCQCSTQRNVRRPESPSKSQSDSVVRRSEGTRAGLSSTPATV EGTPREAKRQGSRGSEARCEAADGEPRKRGGIRAREADEPGKEEEEVSADAASEKPVDGT PGGVSSPEGRAGQGTQEPGKKCGVRGDTSHASREAGTWRLEERENHQTEGSSPPLPWQSE QKDLERSGVGEGDAPPLFSRLASSGLTALEKDVHAGHLEAMESRVALSAAGGRKRRGEEV KERGDSEAHPSEQETLMTSPARLASTGTTRATESTREAPPAGPALGTCSGRGCPSPETAT SGPGKGSLRSCASQDGSPAAIFSSPYKRRSSGRQGRGDEAGSAAPIAGACSRPKDGKATP GVASSSFASGDGCSDGGVDPDRGSAARRRDSSRSLVRRRVSSASPSRPEGPSSGRVSGSR PRHTQSGCSGIVSQGSAFPAALSQWKKGACVSNKCADSDALCCLSPLAYSQQKLLPSHLS GSLAVSEILLESSIVGRYVMLPWNEEDGDIRQNFYVHCPPPVALCSPLSQSVAMHASTPP PFPLGPSRREAVAACPVSPPCGRRGRVNRRSEPERGDGRGRDELESGMYAGAKLAPEGLV KSTQESPVNGRHLASSACGNARAGREEGGRTTEKSRWTKRNYPAPAQGDVTESLESLSYG FGGFPQESRERRRRNTGGGSREACNSRAQGRETRKTEDGAVSSRFEGEARSHPCPRPPAC CTSIPLASASSPTGGSSFAGASVSDPVVRTEAEGEVSPGELGVPGDSAEAPACDLATLSP CGDPGALGRCEEWGPDGKWTDPDGVMSLSRKQQQKFAAWKRLSEIVKDPVVIQDVPNSRA IRQGFVGDCSFLSSLAVLSEFERKHNVPVLSGLLYPQGTIPAVSEAGRRPQDGGERVGPI FNPRGMYACRLYFNGVPRKVLIDDYVPVRKDNKLLAAHSSNKSELWVTLLEKAFVKLMGG SYSMQGSNPGADLYHLTGWIPETIPFRSDVHTGSPASHKPIVTGRENEDDEVVKDPKWTQ IWTQLYLGLTAGRCVACLGTSEVSDAAPSGLDFPEGVSVSSGIVARHAYSILNYAEIEGH RLLYVKNPWGCVRWKGKFSPSDKASWTQNMKEKLGYDPATAAKSDCGCFWIEWLDVVRWF SHLYVCWDNSCFPYQAEIHSKWERSPFIENSSLADDSHMVAFNPQFHLRISPRPEDFASS ALAGPYPFLSFPSLTSPASAASLPMPSNSPPGTPHTASLSPVPSFSQENQPVELWILLSR HVRERQKDLATKYLAIHLHAASERATCPPPPAKQGVYSNGECTLVKLRVHPHVFRDLFAR DASDEGSRFPSDNSAKREEKGKKEAAGGRPPLPGFEGHQLLDASEFVLVVSQYSQKDEFN FTIKVYGHVHSCLTQLPPLLPTDAQSIYFKNSWGPSTAGGCSNDLWRYFTNPHIRLKVPQ PCDAIFFLESPQEHSVNLRLFKNRVATARLLRTGKALSTGPYRAGCCMLKARLEATTYTI IPSTFRPDDFDTFQLSLHVPGNVAKPRPVLLPQPYAIPPPSSLFYRVVDGRKCTRQVWAR VALQVQGPTLVALRLQMPGPPEPGDVFPSLTVYRYTETAEREREVESERSAERKNQKKLQ FVIRSDLDGGLSEDGHTASSAAQDLFQKQGVVNVSSVELVDPLSPYVIFISTLNGDKDAD GCPQYAHSRDVALHYLHRCEGRQREDVPKARPWMLHVIADRPLDVQLLL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_293820 | 208 S | SNSSSPRGA | 0.996 | unsp | TGME49_293820 | 208 S | SNSSSPRGA | 0.996 | unsp | TGME49_293820 | 208 S | SNSSSPRGA | 0.996 | unsp | TGME49_293820 | 213 S | PRGASPSVA | 0.992 | unsp | TGME49_293820 | 237 S | RAAASPIRA | 0.995 | unsp | TGME49_293820 | 258 S | SALPSTREA | 0.99 | unsp | TGME49_293820 | 284 S | LRRHSRFRL | 0.997 | unsp | TGME49_293820 | 404 S | TRRLSPWSP | 0.996 | unsp | TGME49_293820 | 451 S | SSLSSYRSS | 0.994 | unsp | TGME49_293820 | 537 S | ASALSPERR | 0.997 | unsp | TGME49_293820 | 573 S | RRPESPSKS | 0.995 | unsp | TGME49_293820 | 577 S | SPSKSQSDS | 0.997 | unsp | TGME49_293820 | 603 T | TVEGTPREA | 0.994 | unsp | TGME49_293820 | 814 S | RATESTREA | 0.997 | unsp | TGME49_293820 | 870 S | YKRRSSGRQ | 0.997 | unsp | TGME49_293820 | 871 S | KRRSSGRQG | 0.998 | unsp | TGME49_293820 | 931 S | RRRDSSRSL | 0.998 | unsp | TGME49_293820 | 941 S | RRRVSSASP | 0.991 | unsp | TGME49_293820 | 942 S | RRVSSASPS | 0.995 | unsp | TGME49_293820 | 944 S | VSSASPSRP | 0.997 | unsp | TGME49_293820 | 946 S | SASPSRPEG | 0.992 | unsp | TGME49_293820 | 957 S | SGRVSGSRP | 0.997 | unsp | TGME49_293820 | 1111 S | VNRRSEPER | 0.993 | unsp | TGME49_293820 | 1146 S | STQESPVNG | 0.995 | unsp | TGME49_293820 | 1297 S | EGEVSPGEL | 0.992 | unsp | TGME49_293820 | 1537 S | GSPASHKPI | 0.994 | unsp | TGME49_293820 | 1639 S | KGKFSPSDK | 0.993 | unsp | TGME49_293820 | 1731 S | HLRISPRPE | 0.997 | unsp | TGME49_293820 | 1786 S | VPSFSQENQ | 0.993 | unsp | TGME49_293820 | 1863 S | ARDASDEGS | 0.996 | unsp | TGME49_293820 | 1874 S | PSDNSAKRE | 0.994 | unsp | TGME49_293820 | 1914 S | VSQYSQKDE | 0.997 | unsp | TGME49_293820 | 83 S | SSHSSPRST | 0.997 | unsp | TGME49_293820 | 174 S | LDLRSEKED | 0.996 | unsp |