

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
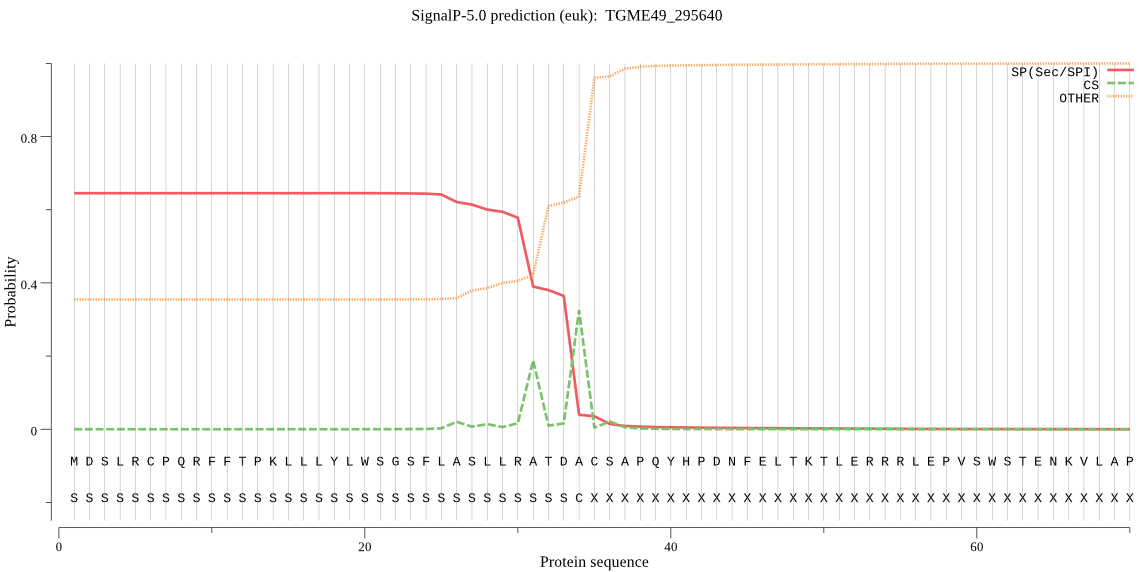
| TGME49_295640 | SP | 0.137530 | 0.860076 | 0.002394 | CS pos: 34-35. TDA-CS. Pr: 0.5810 |
MDSLRCPQRFFTPKLLLYLWSGSFLASLLRATDACSAPQYHPDNFELTKTLERRRLEPVS WSTENKVLAPPKVPGLLAENAAVENFVQSRSISGETADEQNKKHENVLGNLNLDAFLLAA DAGIPAEEQHSGTVNADEHTHGASSSFPRSNASSQTRSYSTLRDESRSTGDTASPVEMNQ AREQHGDEDDASKFSFPFLKPPGRYPPPENLACVDSAQCARIAEMIKRDMNPRADPCYDA EEYFCGGWKTRTSLPADESSWTLSFDTLARDTRDYLKTTLEARSCSDMRDELHRQTPQDS VFSNLNSPSSVIGETGNRVDEWDVLATCMYEACMDEEAIDARGATPLTDRMFGRSERGSM SLEFLLDTDFEAEAIHLAERGNTHPDSSAISTFPVSREAVLVQRLQAIFHRLKDVLFWSA YVGPNELHPDQGQTLNLGSLKLGLSYHFYEEDHLDYQRYYKEHMANILQIFEAHLLKTHP QLLARAQKVNPNLRRSYSERAQRIFDLEKNDLRPLVLSPEETRKVTQYTHEVTFGELKAS TDALDVEALLHLYLDMLPRKLGEQAPSASSLGGKIAFTSSGVVVDDSLVLLIHEKNYFAK LETTLRSVDWDVLHDYILYKVVRSDASMLSRDFRDEIKRYSKQVTKAEPLPRWRTCVSSV PQWILARRYVLGRFDRQKKQTVQLMVTRIRSAFKSLLEEYAWMDEPTREEALEKLAGMQE KIGFPDWLLDDYETYFTRYYGDKSAALELASIHFEVQWHLGLEAIRSQLAEFGEPVDRRE WAMKPHSVNAYFSPSQNEIAFMAAVLQEPSLFVASPHASLGEDTVVKALSYGAIAGVIAH EITHGFDDVGKEYDVKGRLRNWWTPESEEAFKAEATCMKDQYSGYSVVIVDVENDKAIYN TRPSREASEVEHSDGSSGQKTVRVRGDLTLGENIADNGGIQLAWAALKLELSPEQLNSRP LENYGVTLTTAQLFTFSWGHFWCEIALDSFIRRQVETDPHSPARFRIQGPLANFGLFAEQ EQCPVGSLMNPETKCRVW
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_295640 | 168 S | DESRSTGDT | 0.998 | unsp | TGME49_295640 | 168 S | DESRSTGDT | 0.998 | unsp | TGME49_295640 | 168 S | DESRSTGDT | 0.998 | unsp | TGME49_295640 | 284 S | LEARSCSDM | 0.996 | unsp | TGME49_295640 | 359 S | SERGSMSLE | 0.992 | unsp | TGME49_295640 | 496 S | NLRRSYSER | 0.997 | unsp | TGME49_295640 | 498 S | RRSYSERAQ | 0.994 | unsp | TGME49_295640 | 518 S | PLVLSPEET | 0.998 | unsp | TGME49_295640 | 904 S | NTRPSREAS | 0.997 | unsp | TGME49_295640 | 908 S | SREASEVEH | 0.996 | unsp | TGME49_295640 | 1001 S | TDPHSPARF | 0.994 | unsp | TGME49_295640 | 93 S | SRSISGETA | 0.995 | unsp | TGME49_295640 | 166 S | LRDESRSTG | 0.996 | unsp |