

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
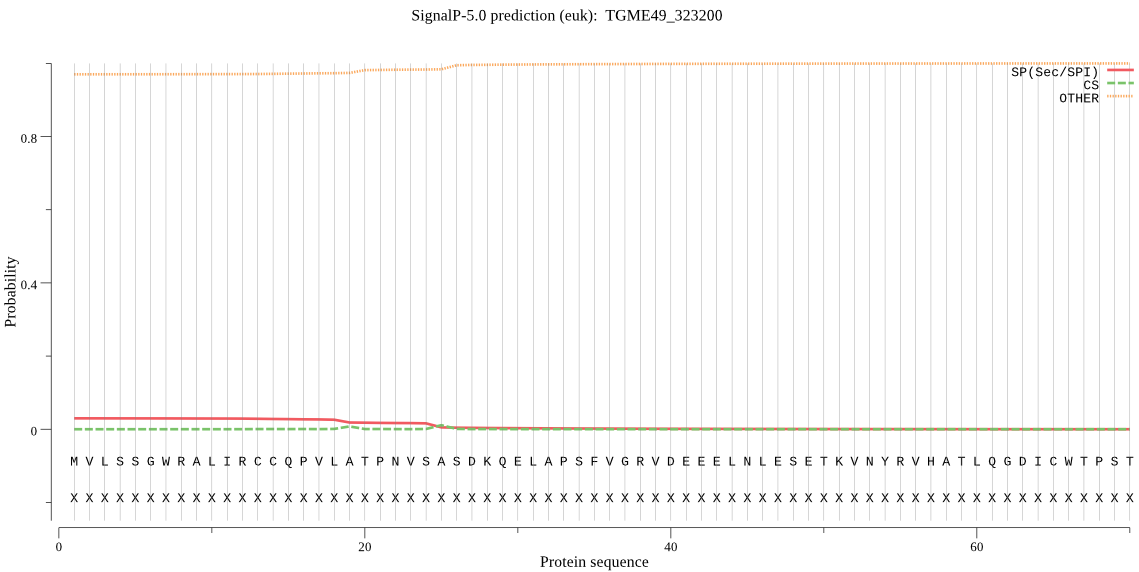
| TGME49_323200 | OTHER | 0.759828 | 0.066744 | 0.173428 |
MVLSSGWRALIRCCQPVLATPNVSASDKQELAPSFVGRVDEEELNLESETKVNYRVHATL QGDICWTPSTASEEFDPESHSCADDAVIVSSFGSRVDAAELDLDRDADIVYDSAYEADNE EDAWSETGSEEDADESPRSSEVLTQADEDGQSFEGDLATTNFVASTQTGEDFSGDEWEIP EEISPAEGMTTRTPTPPGGSPGDCCGDGNEEKIFGRFAALHEKEQPVCQSLSQLPRDMQA PPLEQSVQASKLSQRSSSRPSAKLPPEMLPFIAPHPQWCRRFAYDFKTPAKSLSMVPQPE LSFYDAVVVERHRVAPDGNCQFRSVSYALLGTEDAHAEIRQEVAHYLRGNFNRLSWLINP DTLEEDEGRMARLDKKYRVRIPYKTYKGYPLAEDELKLNWVIRLGDARYRIWGDECTLAV MAEMYNIRIVVEQQEGDGRRATKMGSHAVQVIIPYDVVPEACIPTIFLIYDLQRQHYDVV EKSNPGENVTCPSSTTSCVIQARLVVSHVRTIAKYRRQDCPPSSNQATDCLPRPLFKLRL LSGITTSWA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGME49_323200 | 129 S | SETGSEEDA | 0.996 | unsp | TGME49_323200 | 129 S | SETGSEEDA | 0.996 | unsp | TGME49_323200 | 129 S | SETGSEEDA | 0.996 | unsp | TGME49_323200 | 136 S | DADESPRSS | 0.996 | unsp | TGME49_323200 | 173 S | GEDFSGDEW | 0.992 | unsp | TGME49_323200 | 200 S | PPGGSPGDC | 0.993 | unsp | TGME49_323200 | 257 S | SQRSSSRPS | 0.99 | unsp | TGME49_323200 | 258 S | QRSSSRPSA | 0.995 | unsp | TGME49_323200 | 261 S | SSRPSAKLP | 0.997 | unsp | TGME49_323200 | 302 S | QPELSFYDA | 0.993 | unsp | TGME49_323200 | 72 S | PSTASEEFD | 0.992 | unsp | TGME49_323200 | 113 S | IVYDSAYEA | 0.994 | unsp |