

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg.972.2.1890 | OTHER | 0.898626 | 0.097312 | 0.004062 |
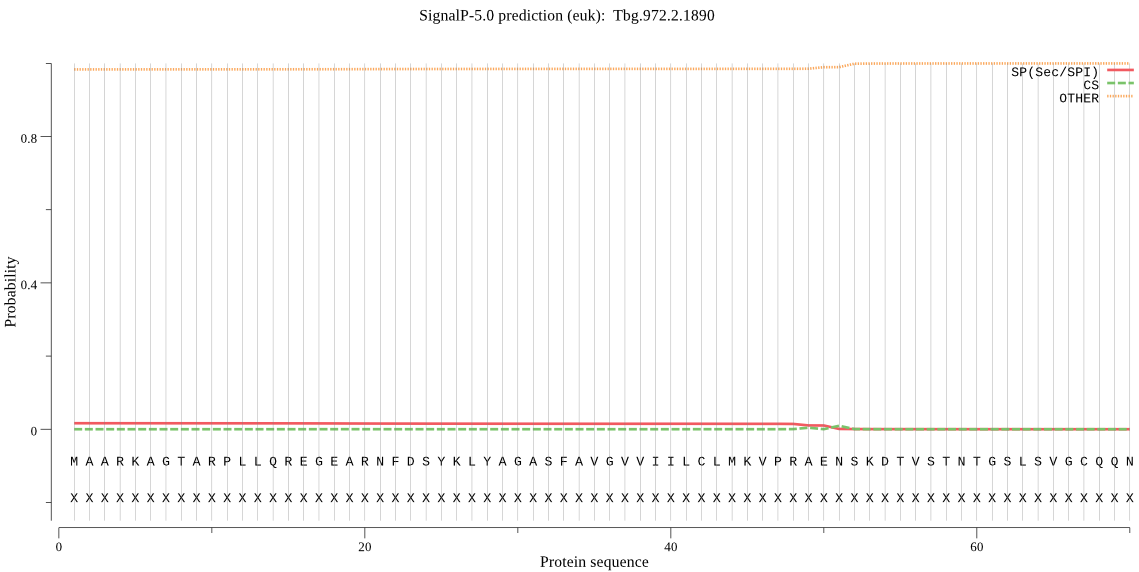
MAARKAGTARPLLQREGEARNFDSYKLYAGASFAVGVVIILCLMKVPRAENSKDTVSTNT GSLSVGCQQNCSAPFGNVLGIYNGVPAMSNCNSDSCTAELWNTVKVEDIRIPAGRVDPHA VPPIYGMQWQCVEYARRYWMLRGTPQPATFGSVDGAADIWDLKDIQLLNGQKRKPLLKYH NGNATSANSKPRVGDLLIYPRQPNGFPYGHVAVVAGVTGDRMFVAEQNWENAAWPGPYHN YSRVLNLSCNPNGTACTVREKDNVTVQGWVRYE