

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg.972.2.1900 | OTHER | 0.999956 | 0.000013 | 0.000031 |
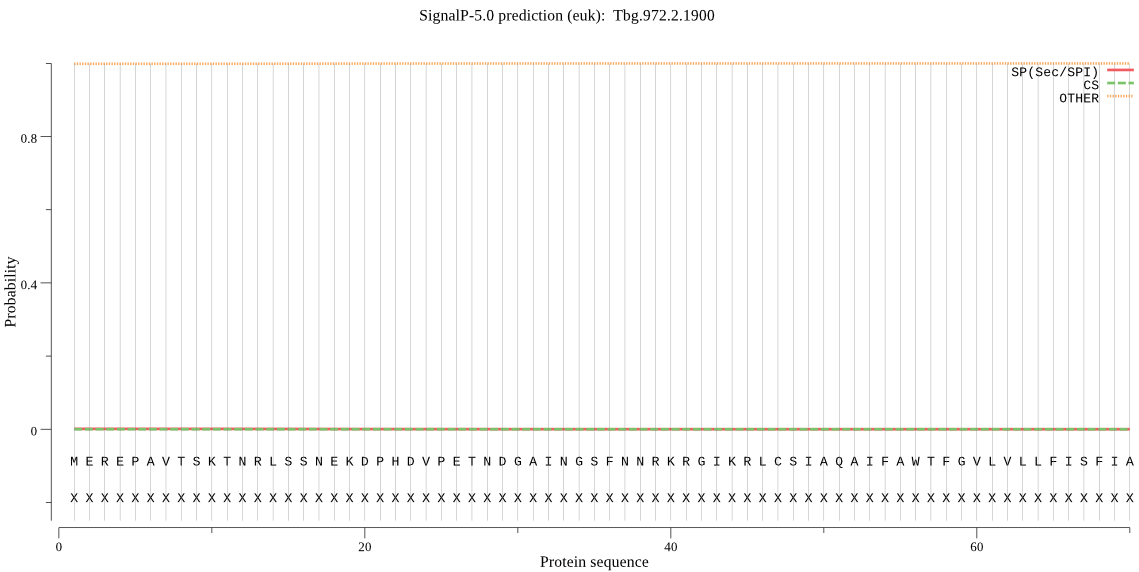
MEREPAVTSKTNRLSSNEKDPHDVPETNDGAINGSFNNRKRGIKRLCSIAQAIFAWTFGV LVLLFISFIAFGVLYSGIRYPEGSGKVEKHCLNPVDAILGAHEGVFSYSNCGATENTTTY NNVTVAGTSYQSGLKWQCVEYARRYWMLRGTPQPATFGSVDGAADIWDLKDIQLLNGQKR KPLLKYHNGNATSANSKPRVGDLLIYPRQPNGFPCGHVAVVVGVTGDRMFVAEQNWENAA WPGPYHNYSRVLNLSCNPNGTACTVREKDNVTVQGWVRYE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Tbg.972.2.1900 | 15 S | TNRLSSNEK | 0.995 | unsp | Tbg.972.2.1900 | 16 S | NRLSSNEKD | 0.997 | unsp |