

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
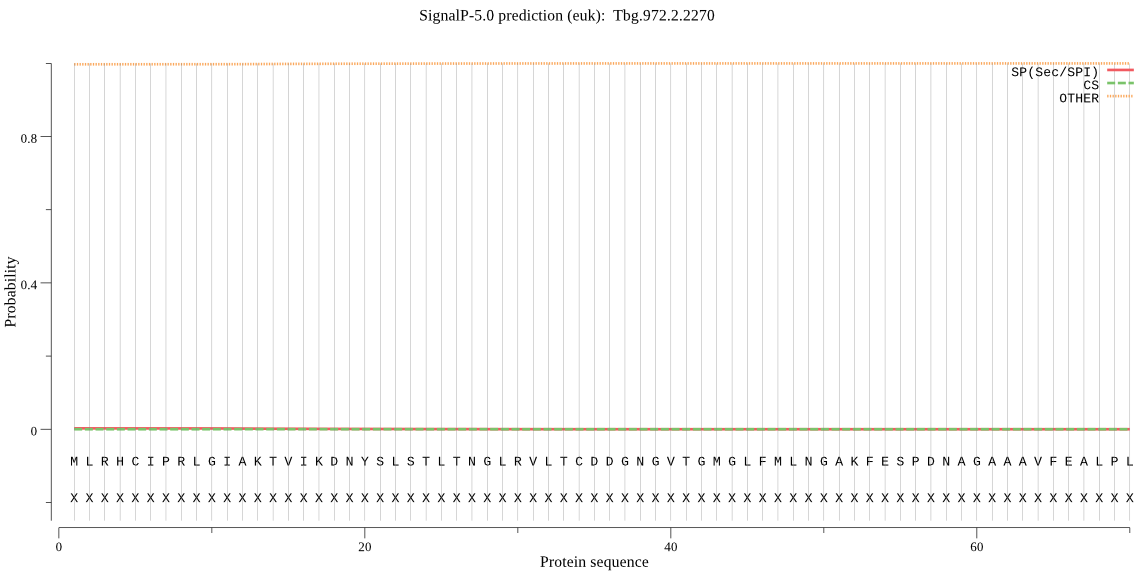
| Tbg.972.2.2270 | OTHER | 0.706224 | 0.001322 | 0.292453 |
MLRHCIPRLGIAKTVIKDNYSLSTLTNGLRVLTCDDGNGVTGMGLFMLNGAKFESPDNAG AAAVFEALPLRDNQIYTSREISQALSGLGNAFKVTNNKEALSVILMLPRYHQRDGLELLN AMCLHPTRNEEEFRVAKEKTHERTLLYDRDATSVCFELVHEAGWNGKGLGHSLNPKKEEL DKLTLEKFTAFHSACTRPERTVLAATGVADHKSFAEEVEKLLRFNNADVAVQAMPQLQPG YYPYTGGSRLVHRTEAPESVNKFQEKSLSHVALFFQGVPINHPDYYNISVIQTLLGGGTS FSSGGPGKGMQTKLFREVLNREGFLHGLECITAWYSDGGLFGLYGTAPHQAVVSLLNVMI YQAASICQRVSPTHLEMAKNQLRSQLILLGEGREQLLSDMGFNLVVHNHIITATETMEGT RNITLDDLKRVCADMIKKPLTFTVYGDTTKMPSHESLEETVRKTYEKLN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tbg.972.2.2270 | 456 S | PSHESLEET | 0.996 | unsp | Tbg.972.2.2270 | 456 S | PSHESLEET | 0.996 | unsp | Tbg.972.2.2270 | 456 S | PSHESLEET | 0.996 | unsp | Tbg.972.2.2270 | 371 S | CQRVSPTHL | 0.994 | unsp | Tbg.972.2.2270 | 453 S | TKMPSHESL | 0.996 | unsp |