

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg972.1.1810 | OTHER | 0.993724 | 0.002334 | 0.003942 |
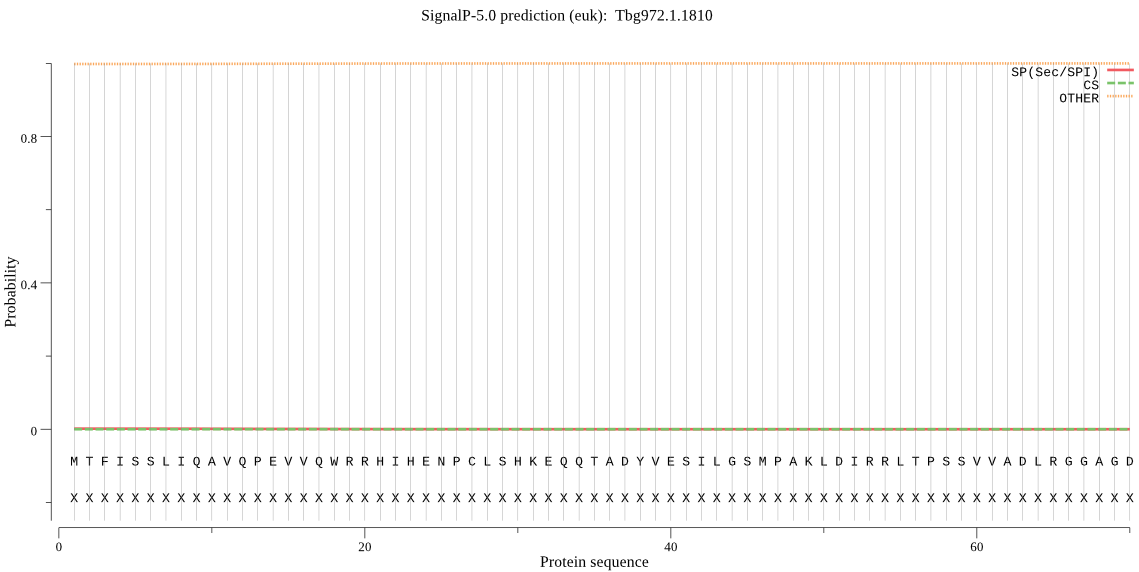
MTFISSLIQAVQPEVVQWRRHIHENPCLSHKEQQTADYVESILGSMPAKLDIRRLTPSSV VADLRGGAGDGPTYALRADMDALPLQEESGEPFSSKLPGVMHACGHDAHTAILLGAIKVL CHVKDKICGTIRFIFQHAEEVIPSGAKQLVQLGVLDGVKMIFGLAVDVSNPPGTVLCKSG VLTSACNDFDIVIQGASGHASQPELCIDPIVIGAQVVMSLQTIVSRRIGALTTPVLSIAT FQGGRGGYNVIPDTVHLRGTLRCLDSGVQKRVPVMVEEIVAGITSAHGAKHTVSWLEPNI VTYNNEAAYDVVKRVAEHVVSGITFVELPTAMTGVTDFGEYGAVIPGCLFLLGAGNETNN SVSNHNYSSRFRLNENVMVDGVRMFVGLMMTLAVPDAPSPGT
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.1.1810 | 29 S | NPCLSHKEQ | 0.994 | unsp | Tbg972.1.1810 | 56 T | IRRLTPSSV | 0.993 | unsp |