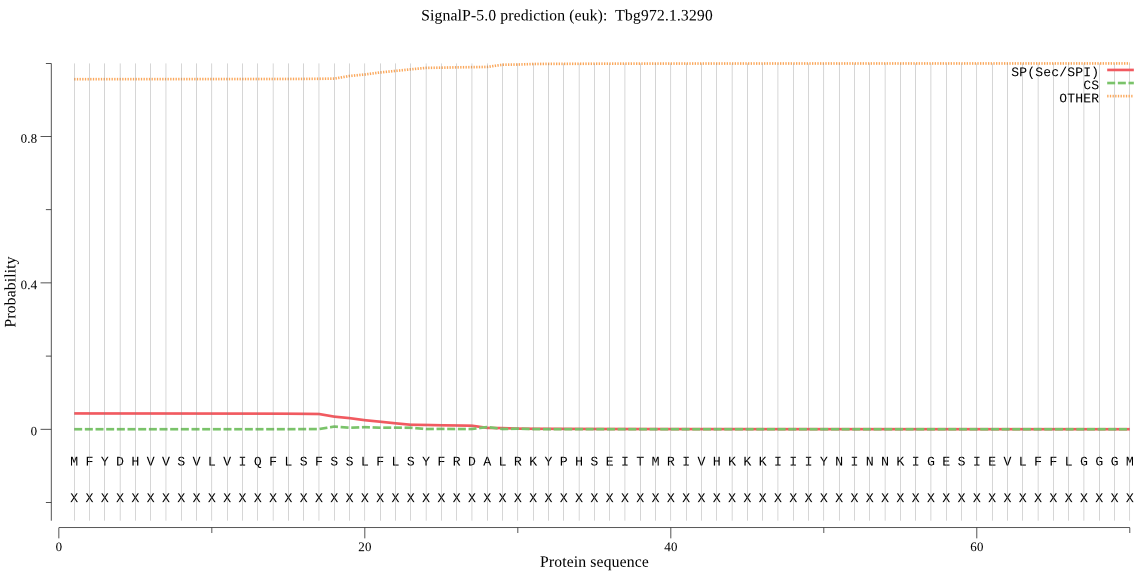
Fasta :-
MFYDHVVSVLVIQFLSFSSLFLSYFRDALRKYPHSEITMRIVHKKKIIIYNINNKIGESI
EVLFFLGGGMAAFFGSLVKSFILPKPSPTYSADKHPGKLVHIPRVDWDTRKENGTFTYGL
LLLDTAAKFIIIYAHTNAVDVAMVFETMSYVSKRTSTSVLLVEYTGYGIAYGETTERSMN
EDVLSAYYYAVRHMRVPADRVVLMGRSIGTGPSAQVCALLQGEEEVPALLVLQSPFTSLK
ECANDITPNVGSIVGYLGYDWFRTIDVVAQVRCPIIIHHGQCDDVVPFEHAQQLKRTIEE
ATPPGVVELHAEPNRGHNDLPTESANRFIDKKLRSFGQPRCLQPRCRPYHLVNPSIYEYL
FCVKEQPIINMEELLKHWNETLSIGSFAYKREKLYVLLTASVSLFAMRCARAWQHYAGSR
KRHYGNSYIASCNGSVGVGVGVGVVGGSGSGGNGDGSGSGAEELYCAKEDIIKRCLACWG
SPLGAYLSVRGPPIRHKIFGVHLDVCVENVVASDGISNESGFRSCGGNPYFLKRDTGEAY
LSVAELEFTHGLTRSVAAAMSTAPTLGGEEDEMDVFLQKNVVTRIQTQCERVVAFLDSNE
WENMLEVLVSFGQRAPSFLSSKALQYYSECSSQARFGDSCGTGAACCGNDSGPAPRSEVM
ETIEDVDEWLRPWVVSPDTRMQLGTEVPWDYYLLKARLCVVEYTPLGPDTSWEEARRITD
AWRVVKIIHDLFCSYSRRVLHPSFSAS