

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
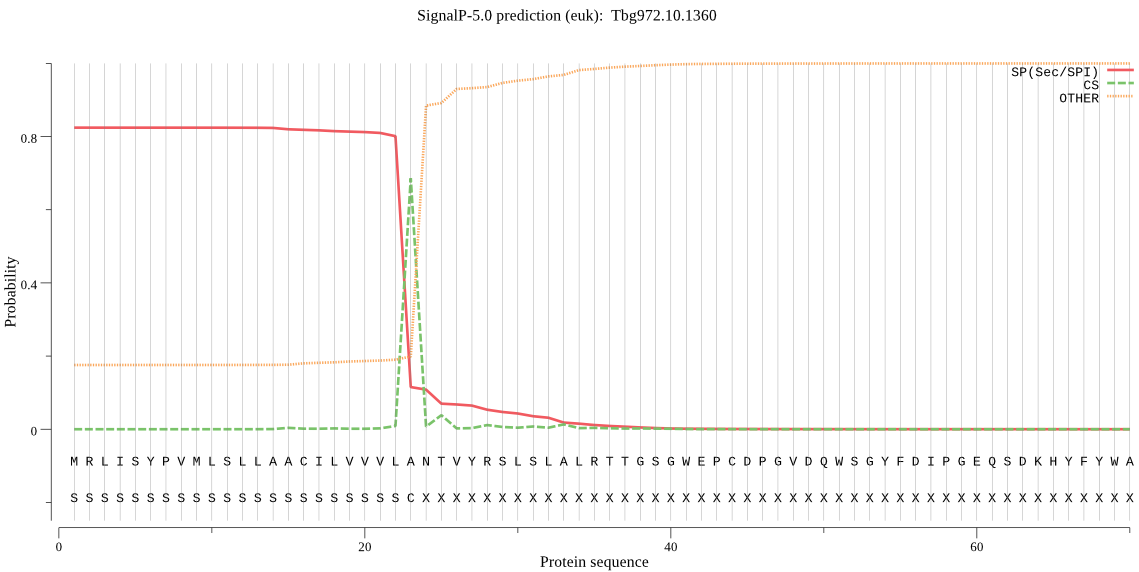
| Tbg972.10.1360 | SP | 0.044650 | 0.953836 | 0.001514 | CS pos: 23-24. VLA-NT. Pr: 0.9117 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.10.1360 | 298 S | LTKISTYDI | 0.996 | unsp | Tbg972.10.1360 | 410 S | DTPFSSIDG | 0.995 | unsp |