

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| Tbg972.10.1400 | SP | 0.000334 | 0.999585 | 0.000080 | CS pos: 21-22. ASA-LQ. Pr: 0.7869 |
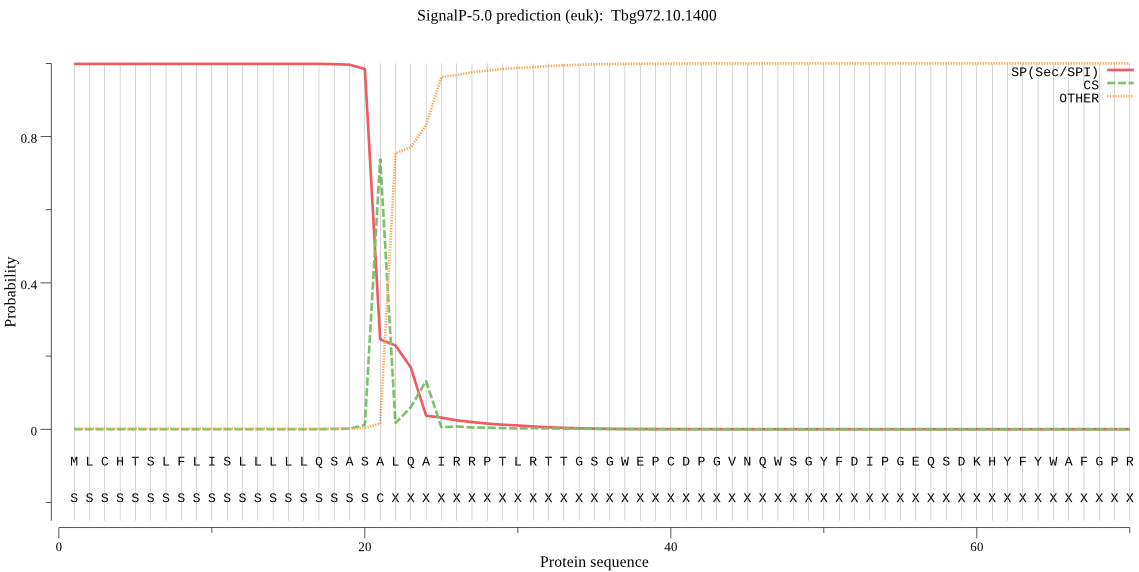
MLCHTSLFLISLLLLLQSASALQAIRRPTLRTTGSGWEPCDPGVNQWSGYFDIPGEQSDK HYFYWAFGPRDGNPNAPVLLWMTGGPGCSSMFALLAENGPCLMNETTGDIYNNTYSWNNH AYVIYIDQPAGVGFSYADKADYDKNEAEVSEDMYNFLQAFFGEHEDLRENDFFVVGESYG GHFAPATAYRINQGNKKGEGIYIPLAGLAVGNGLTDPYTQYASYPRLAWDWCKEVLGYSC ISRETYDSMNSMVPACQSNISACDADNSSSADSYCEMAGAACSGFVSDFLLTGINVYDIR KTCDGPLCYNTTGVDNFMNREDVQRSLGVDPMTWQACNMEVNEMFDIDWFKNFNYTISGL LEDGVRVMIYAGDMDFICNWIGNKEWTLALQWSGSEEFVKAPDTPFSSIDGSAAGLVRSV SSNTSSMHFSFVQVYRAGHMVPMDQPAAASTIIEKFMRNEPLS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tbg972.10.1400 | 407 S | DTPFSSIDG | 0.995 | unsp | Tbg972.10.1400 | 407 S | DTPFSSIDG | 0.995 | unsp | Tbg972.10.1400 | 407 S | DTPFSSIDG | 0.995 | unsp | Tbg972.10.1400 | 29 T | IRRPTLRTT | 0.992 | unsp | Tbg972.10.1400 | 273 S | SSADSYCEM | 0.996 | unsp |