

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
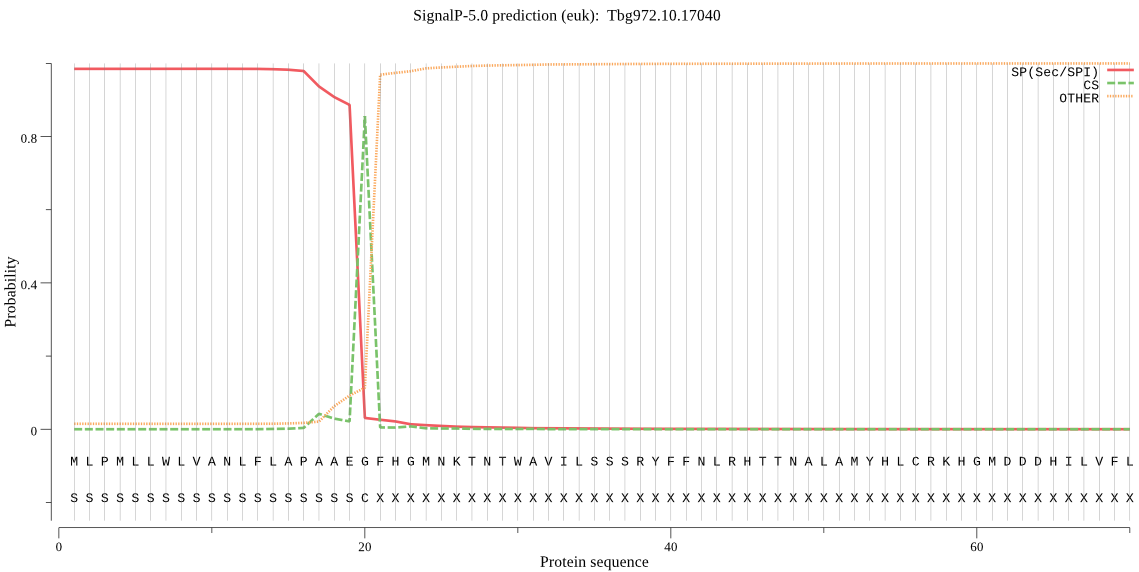
| Tbg972.10.17040 | SP | 0.000803 | 0.999177 | 0.000020 | CS pos: 20-21. AEG-FH. Pr: 0.9489 |
MLPMLLWLVANLFLAPAAEGFHGMNKTNTWAVILSSSRYFFNLRHTTNALAMYHLCRKHG MDDDHILVFLSDSYACDPRKPNPATIYGAPAQAEQPNLYGCNIRVDYASYDVGVRRFLGV LQGRYDENTPPSRRLDTDENSNIIIYAAGHSAEKFFKFQDSEFMSSTDIADTLMMMWEQR RYRKLVFLVDTCRALSLCLEIKAPNVVCLASSEAHLDSYSHHLDPPSGFTVITRWTFEFL EVLKDSKCRPENGEVTLLQKSFYDFNYGPERLSLPQPLSEPAHFDAVNRPNAIREWKMDE FFCEQDRDKIPVELRYDLF